Extracellular vesicles secreted by human aneuploid embryos present a distinct transcriptomic profile and upregulate MUC1 transcription in decidualised endometrial stromal cells
- PMID: 38559895
- PMCID: PMC10980593
- DOI: 10.1093/hropen/hoae014
Extracellular vesicles secreted by human aneuploid embryos present a distinct transcriptomic profile and upregulate MUC1 transcription in decidualised endometrial stromal cells
Abstract
Study question: Do extracellular vesicles (EVs) secreted by aneuploid human embryos possess a unique transcriptomic profile that elicits a relevant transcriptomic response in decidualized primary endometrial stromal cells (dESCs)?
Summary answer: Aneuploid embryo-derived EVs contain transcripts of PPM1J, LINC00561, ANKRD34C, and TMED10 with differential abundance from euploid embryo-derived EVs and induce upregulation of MUC1 transcript in dESCs.
What is known already: We have previously reported that IVF embryos secrete EVs that can be internalized by ESCs, conceptualizing that successful implantation to the endometrium is facilitated by EVs. Whether these EVs may additionally serve as biomarkers of ploidy status is unknown.
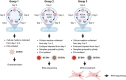
Study design size duration: Embryos destined for biopsy for preimplantation genetic testing for aneuploidy (PGT-A) were grown under standard conditions. Spent media (30 μl) were collected from euploid (n = 175) and aneuploid (n = 140) embryos at cleavage (Days 1-3) stage and from euploid (n = 187) and aneuploid (n = 142) embryos at blastocyst (Days 3-5) stage. Media samples from n = 35 cleavage-stage embryos were pooled in order to obtain five euploid and four aneuploid pools. Similarly, media samples from blastocysts were pooled to create one euploid and one aneuploid pool. ESCs were obtained from five women undergoing diagnostic laparoscopy.
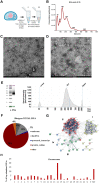
Participants/materials setting methods: EVs were isolated from pools of media by differential centrifugation and EV-RNA sequencing was performed following a single-cell approach that circumvents RNA extraction. ESCs were decidualized (estradiol: 10 nM, progesterone: 1 µM, cAMP: 0.5 mM twice every 48 h) and incubated for 24 h with EVs (50 ng/ml). RNA sequencing was performed on ESCs.
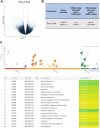
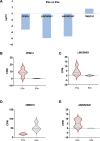
Main results and the role of chance: Aneuploid cleavage stage embryos secreted EVs that were less abundant in RNA fragments originating from the genes PPM1J (log2fc = -5.13, P = 0.011), LINC00561 (log2fc = -7.87, P = 0.010), and ANKRD34C (log2fc = -7.30, P = 0.017) and more abundant in TMED10 (log2fc = 1.63, P = 0.025) compared to EVs of euploid embryos. Decidualization per se induced downregulation of MUC1 (log2fc = -0.54, P = 0.0028) in ESCs as a prerequisite for the establishment of receptive endometrium. The expression of MUC1 transcript in decidualized ESCs was significantly increased following treatment with aneuploid compared to euploid embryo-secreted EVs (log2fc = 0.85, P = 0.0201).
Large scale data: Raw data have been uploaded to GEO (accession number GSE234338).
Limitations reasons for caution: The findings of the study will require validation utilizing a second cohort of EV samples.
Wider implications of the findings: The discovery that the transcriptomic profile of EVs secreted from aneuploid cleavage stage embryos differs from that of euploid embryos supports the possibility to develop a non-invasive methodology for PGT-A. The upregulation of MUC1 in dESCs following aneuploid embryo EV treatment proposes a new mechanism underlying implantation failure.
Study funding/competing interests: The study was supported by a Marie Skłodowska-Curie Actions fellowship awarded to SM by the European Commission (CERVINO grant agreement ID: 79620) and by a BIRTH research grant from Theramex HQ UK Ltd. The authors have no conflicts of interest to declare.
Keywords: aneuploidy; embryo; endometrium; extracellular vesicles; implantation; non-invasive preimplantation genetic testing for aneuploidy.
© The Author(s) 2024. Published by Oxford University Press on behalf of European Society of Human Reproduction and Embryology.
Conflict of interest statement
None to declare.
Figures



References
-
- Aleksejeva E, Zarovni N, Dissanayake K, Godakumara K, Vigano P, Fazeli A, Jaakma Ü, Salumets A.. Extracellular vesicle research in reproductive science: Paving the way for clinical achievements†. Biol Reprod 2022;106:408–424. - PubMed
-
- Cimadomo D, Rienzi L, Conforti A, Forman E, Canosa S, Innocenti F, Poli M, Hynes J, Gemmell L, Vaiarelli A. et al. Opening the black box: why do euploid blastocysts fail to implant? A systematic review and meta-analysis. Hum Reprod Update 2023;29:570–633. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
