CircGPC3 promotes hepatocellular carcinoma progression and metastasis by sponging miR-578 and regulating RAB7A/PSME3 expression
- PMID: 38561366
- PMCID: PMC10984923
- DOI: 10.1038/s41598-024-58004-y
CircGPC3 promotes hepatocellular carcinoma progression and metastasis by sponging miR-578 and regulating RAB7A/PSME3 expression
Abstract
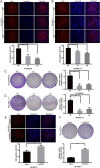
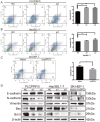
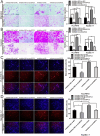
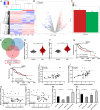
CircRNAs are a class of highly stable noncoding RNAs that play an important role in the progression of many diseases, especially cancer. In this study, high-throughput sequencing was used to screen for abnormally expressed circRNAs, and we found that circGPC3 was overexpressed in HCC tissues. However, the underlying mechanism of circGPC3 in the development and metastasis of hepatocellular carcinoma (HCC) remains unknown. In our study, we found that circGPC3 was significantly upregulated in HCC tissues and cells and that its overexpression was positively correlated with overall survival, TNM stage and lymph node metastasis. In vivo and in vitro experiments showed that circGPC3 knockdown repressed HCC cell migration, invasion and proliferation and promoted apoptosis. Mechanistically, circGPC3 promoted HCC proliferation and metastasis through the miR-578/RAB7A/PSME3 axis. Our results demonstrate that circGPC3 contributes to the progression of HCC and provides an intervention target for HCC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Long non-coding RNA PTTG3P functions as an oncogene by sponging miR-383 and up-regulating CCND1 and PARP2 in hepatocellular carcinoma.BMC Cancer. 2019 Jul 24;19(1):731. doi: 10.1186/s12885-019-5936-2. BMC Cancer. 2019. PMID: 31340767 Free PMC article.
-
Long noncoding RNA GIHCG promotes hepatocellular carcinoma progression through epigenetically regulating miR-200b/a/429.J Mol Med (Berl). 2016 Nov;94(11):1281-1296. doi: 10.1007/s00109-016-1442-z. Epub 2016 Jul 5. J Mol Med (Berl). 2016. PMID: 27380494
-
circSLCO1B7 suppresses the malignant progression of hepatocellular carcinoma via the miR-556-3p/DAB2IP axis.Aging (Albany NY). 2023 Nov 24;15(22):13329-13344. doi: 10.18632/aging.205244. Epub 2023 Nov 24. Aging (Albany NY). 2023. PMID: 38015711 Free PMC article.
-
Long Noncoding RNA HOTAIR Contributes to Progression in Hepatocellular Carcinoma by Sponging miR-217-5p.Cancer Biother Radiopharm. 2020 Jun;35(5):387-396. doi: 10.1089/cbr.2019.3070. Epub 2020 Apr 21. Cancer Biother Radiopharm. 2020. Retraction in: Cancer Biother Radiopharm. 2021 Sep;36(7):613. doi: 10.1089/cbr.2019.3070.retract. PMID: 32315535 Retracted.
-
Elevated expression of circular RNA circ_0008450 predicts dismal prognosis in hepatocellular carcinoma and regulates cell proliferation, apoptosis, and invasion via sponging miR-548p.J Cell Biochem. 2019 Jun;120(6):9487-9494. doi: 10.1002/jcb.28224. Epub 2018 Dec 16. J Cell Biochem. 2019. PMID: 30556306
Cited by
-
Expression Profile of Serum CircFUNDC1 and CircUHRF1 Can Differentiate Between Colorectal Cancer and Inflammatory Bowel Diseases (Ulcerative Colitis and Crohn's Disease).J Clin Lab Anal. 2025 Jun;39(11):e70039. doi: 10.1002/jcla.70039. Epub 2025 May 16. J Clin Lab Anal. 2025. PMID: 40376950 Free PMC article.
-
circ-1584 selectively promotes the antitumor activity of the oncolytic virus M1 on pancreatic cancer.Mol Ther Oncol. 2024 Dec 17;33(1):200919. doi: 10.1016/j.omton.2024.200919. eCollection 2025 Mar 20. Mol Ther Oncol. 2024. PMID: 39866243 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

