The miR-183/96/182 cluster regulates sensory innervation, resident myeloid cells and functions of the cornea through cell type-specific target genes
- PMID: 38561433
- PMCID: PMC10985120
- DOI: 10.1038/s41598-024-58403-1
The miR-183/96/182 cluster regulates sensory innervation, resident myeloid cells and functions of the cornea through cell type-specific target genes
Erratum in
-
Correction: The miR-183/96/182 cluster regulates sensory innervation, resident myeloid cells and functions of the cornea through cell type-specific target genes.Sci Rep. 2025 Jun 25;15(1):20297. doi: 10.1038/s41598-025-08233-6. Sci Rep. 2025. PMID: 40562818 Free PMC article. No abstract available.
Abstract
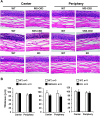
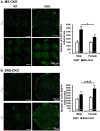
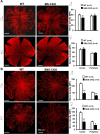
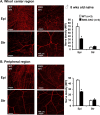
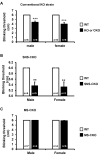
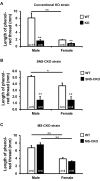
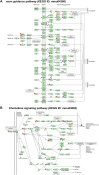
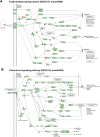
The conserved miR-183/96/182 cluster (miR-183C) is expressed in both corneal resident myeloid cells (CRMCs) and sensory nerves (CSN) and modulates corneal immune/inflammatory responses. To uncover cell type-specific roles of miR-183C in CRMC and CSN and their contributions to corneal physiology, myeloid-specific miR-183C conditional knockout (MS-CKO), and sensory nerve-specific CKO (SNS-CKO) mice were produced and characterized in comparison to the conventional miR-183C KO. Immunofluorescence and confocal microscopy of flatmount corneas, corneal sensitivity, and tear volume assays were performed in young adult naïve mice; 3' RNA sequencing (Seq) and proteomics in the trigeminal ganglion (TG), cornea and CRMCs. Our results showed that, similar to conventional KO mice, the numbers of CRMCs were increased in both MS-CKO and SNS-CKO vs age- and sex-matched WT control littermates, suggesting intrinsic and extrinsic regulations of miR-183C on CRMCs. The number of CRMCs was increased in male vs female MS-CKO mice, suggesting sex-dependent regulation of miR-183C on CRMCs. In the miR-183C KO and SNS-CKO, but not the MS-CKO mice, CSN density was decreased in the epithelial layer of the cornea, but not the stromal layer. Functionally, corneal sensitivity and basal tear volume were reduced in the KO and SNS-CKO, but not the MS-CKO mice. Tear volume in males is consistently higher than female WT mice. Bioinformatic analyses of the transcriptomes revealed a series of cell-type specific target genes of miR-183C in TG sensory neurons and CRMCs. Our data elucidate that miR-183C imposes intrinsic and extrinsic regulation on the establishment and function of CSN and CRMCs by cell-specific target genes. miR-183C modulates corneal sensitivity and tear production through its regulation of corneal sensory innervation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
New insight into the neuroimmune interplay in Pseudomonas aeruginosa keratitis.Ocul Surf. 2025 Jul 24;38:170-183. doi: 10.1016/j.jtos.2025.07.008. Online ahead of print. Ocul Surf. 2025. PMID: 40714297
-
New Insight Into the Neuroimmune Interplay In Pseudomonas aeruginosa Keratitis.bioRxiv [Preprint]. 2025 Mar 11:2025.03.06.641908. doi: 10.1101/2025.03.06.641908. bioRxiv. 2025. Update in: Ocul Surf. 2025 Jul 24;38:170-183. doi: 10.1016/j.jtos.2025.07.008. PMID: 40161776 Free PMC article. Updated. Preprint.
-
The miR-183/96/182 cluster is a checkpoint for resident immune cells and shapes the cellular landscape of the cornea.Ocul Surf. 2023 Oct;30:17-41. doi: 10.1016/j.jtos.2023.07.012. Epub 2023 Aug 2. Ocul Surf. 2023. PMID: 37536656 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Artificial intelligence for detecting keratoconus.Cochrane Database Syst Rev. 2023 Nov 15;11(11):CD014911. doi: 10.1002/14651858.CD014911.pub2. Cochrane Database Syst Rev. 2023. PMID: 37965960 Free PMC article.
Cited by
-
New insight into the neuroimmune interplay in Pseudomonas aeruginosa keratitis.Ocul Surf. 2025 Jul 24;38:170-183. doi: 10.1016/j.jtos.2025.07.008. Online ahead of print. Ocul Surf. 2025. PMID: 40714297
-
New Insight Into the Neuroimmune Interplay In Pseudomonas aeruginosa Keratitis.bioRxiv [Preprint]. 2025 Mar 11:2025.03.06.641908. doi: 10.1101/2025.03.06.641908. bioRxiv. 2025. Update in: Ocul Surf. 2025 Jul 24;38:170-183. doi: 10.1016/j.jtos.2025.07.008. PMID: 40161776 Free PMC article. Updated. Preprint.
References
-
- Lwigale, P. Y. Corneal development: Different cells from a common progenitor. Prog. Mol. Biol. Transl. Sci.134, 43–59. 10.1016/bs.pmbts.2015.04.003 (2015). - PubMed
-
- DelMonte, D. W. & Kim, T. Anatomy and physiology of the cornea. J. Cataract. Refract. Surg.37, 588–598. 10.1016/j.jcrs.2010.12.037 (2011). - PubMed
-
- Sosnova, M., Bradl, M. & Forrester, J. V. CD34+ corneal stromal cells are bone marrow-derived and express hemopoietic stem cell markers. Stem Cells23, 507–515. 10.1634/stemcells.2004-0291 (2005). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

