Detection and classification of the integrative conjugative elements of Lactococcus lactis
- PMID: 38561675
- PMCID: PMC10983677
- DOI: 10.1186/s12864-024-10255-9
Detection and classification of the integrative conjugative elements of Lactococcus lactis
Abstract
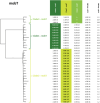
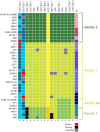
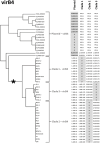
Lactococcus lactis is widely applied by the dairy industry for the fermentation of milk into products such as cheese. Adaptation of L. lactis to the dairy environment often depends on functions encoded by mobile genetic elements (MGEs) such as plasmids. Other L. lactis MGEs that contribute to industrially relevant traits like antimicrobial production and carbohydrate utilization capacities belong to the integrative conjugative elements (ICE). Here we investigate the prevalence of ICEs in L. lactis using an automated search engine that detects colocalized, ICE-associated core-functions (involved in conjugation or mobilization) in lactococcal genomes. This approach enabled the detection of 36 candidate-ICEs in 69 L. lactis genomes. By phylogenetic analysis of conserved protein functions encoded in all lactococcal ICEs, these 36 ICEs could be classified in three main ICE-families that encompass 7 distinguishable ICE-integrases and are characterized by apparent modular-exchangeability and plasticity. Finally, we demonstrate that phylogenetic analysis of the conjugation-associated VirB4 ATPase function differentiates ICE- and plasmid-derived conjugation systems, indicating that conjugal transfer of lactococcal ICEs and plasmids involves genetically distinct machineries. Our genomic analysis and sequence-based classification of lactococcal ICEs creates a comprehensive overview of the conserved functional repertoires encoded by this family of MGEs in L. lactis, which can facilitate the future exploitation of the functional traits they encode by ICE mobilization to appropriate starter culture strains.
Keywords: Lactococcus lactis; Comparative genomics; Integrative conjugative element (ICE); Mobile genetic elements.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

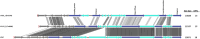

Similar articles
-
The lactococcal ICE-ome encodes a repertoire of exchangeable traits with potential industrial relevance.BMC Genomics. 2024 Jul 30;25(1):734. doi: 10.1186/s12864-024-10646-y. BMC Genomics. 2024. PMID: 39080539 Free PMC article.
-
Versatile Cas9-Driven Subpopulation Selection Toolbox for Lactococcus lactis.Appl Environ Microbiol. 2018 Apr 2;84(8):e02752-17. doi: 10.1128/AEM.02752-17. Print 2018 Apr 15. Appl Environ Microbiol. 2018. PMID: 29453254 Free PMC article.
-
The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation.PLoS Genet. 2011 Aug;7(8):e1002222. doi: 10.1371/journal.pgen.1002222. Epub 2011 Aug 18. PLoS Genet. 2011. PMID: 21876676 Free PMC article.
-
The Role of Integrative and Conjugative Elements in Antibiotic Resistance Evolution.Trends Microbiol. 2021 Jan;29(1):8-18. doi: 10.1016/j.tim.2020.05.011. Epub 2020 Jun 11. Trends Microbiol. 2021. PMID: 32536522 Review.
-
The Lactococcus lactis plasmidome: much learnt, yet still lots to discover.FEMS Microbiol Rev. 2014 Sep;38(5):1066-88. doi: 10.1111/1574-6976.12074. Epub 2014 Jun 23. FEMS Microbiol Rev. 2014. PMID: 24861818 Review.
Cited by
-
The lactococcal ICE-ome encodes a repertoire of exchangeable traits with potential industrial relevance.BMC Genomics. 2024 Jul 30;25(1):734. doi: 10.1186/s12864-024-10646-y. BMC Genomics. 2024. PMID: 39080539 Free PMC article.
-
Comprehensive profiling of integrative conjugative elements (ICEs) in Mollicutes: distinct catalysts of gene flow and genome shaping.NAR Genom Bioinform. 2025 Jun 23;7(2):lqaf083. doi: 10.1093/nargab/lqaf083. eCollection 2025 Jun. NAR Genom Bioinform. 2025. PMID: 40585299 Free PMC article.
References
-
- Andersen JM, Pedersen CM, Bang-Berthelsen CH. Omics-based comparative analysis of putative mobile genetic elements in Lactococcus lactis. In: FEMS microbiology letters, vol. 366. Oxford Academic; 2019. p. 102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

