Circulating cell-free DNA methylation-based multi-omics analysis allows early diagnosis of pancreatic ductal adenocarcinoma
- PMID: 38561976
- PMCID: PMC11547243
- DOI: 10.1002/1878-0261.13643
Circulating cell-free DNA methylation-based multi-omics analysis allows early diagnosis of pancreatic ductal adenocarcinoma
Abstract
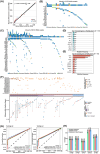
Pancreatic ductal adenocarcinoma (PDAC) is a highly aggressive cancer with a 5-year survival rate of 7.2% in China. However, effective approaches for diagnosis of PDAC are limited. Tumor-originating genomic and epigenomic aberration in circulating free DNA (cfDNA) have potential as liquid biopsy biomarkers for cancer diagnosis. Our study aims to assess the feasibility of cfDNA-based liquid biopsy assay for PDAC diagnosis. In this study, we performed parallel genomic and epigenomic profiling of plasma cfDNA from Chinese PDAC patients and healthy individuals. Diagnostic models were built to distinguish PDAC patients from healthy individuals. Cancer-specific changes in cfDNA methylation landscape were identified, and a diagnostic model based on six methylation markers achieved high sensitivity (88.7% for overall cases and 78.0% for stage I patients) and specificity (96.8%), outperforming the mutation-based model significantly. Moreover, the combination of the methylation-based model with carbohydrate antigen 19-9 (CA19-9) levels further improved the performance (sensitivity: 95.7% for overall cases and 95.5% for stage I patients; specificity: 93.3%). In conclusion, our findings suggest that both methylation-based and integrated liquid biopsy assays hold promise as non-invasive tools for detection of PDAC.
Keywords: cfDNA; liquid biopsy; machine learning; methylation; mutation; pancreatic ductal adenocarcinoma.
© 2024 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
GZ, SG, WD, YZ, WW, TK, YR, JY, GJ and WL have declared no competing interest. RJ, YS, ZL, JS, JP, and YW are employees of Envelope Health Biotechnology Co. Ltd., BGI‐Shenzhen. SZ is an employee of BGI Genomics, BGI‐Shenzhen.
Figures




Similar articles
-
Novel mRNA biomarker-based liquid biopsy for the detection of resectable pancreatic cancer.BMC Cancer. 2025 Apr 23;25(1):762. doi: 10.1186/s12885-025-14124-w. BMC Cancer. 2025. PMID: 40269781 Free PMC article.
-
Plasma and Urine Circulating Tumor DNA Methylation Profiles for Non-Invasive Pancreatic Ductal Adenocarcinoma Detection: Significant Findings in Plasma Only.Int J Mol Sci. 2025 May 22;26(11):4972. doi: 10.3390/ijms26114972. Int J Mol Sci. 2025. PMID: 40507783 Free PMC article.
-
High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients.Ann Oncol. 2017 Apr 1;28(4):741-747. doi: 10.1093/annonc/mdx004. Ann Oncol. 2017. PMID: 28104621 Free PMC article.
-
Circulating Tumor Cells and Cell-Free DNA in Pancreatic Ductal Adenocarcinoma.Am J Pathol. 2019 Jan;189(1):71-81. doi: 10.1016/j.ajpath.2018.03.020. Am J Pathol. 2019. PMID: 30558725 Review.
-
Cell-free DNA: plays an essential role in early diagnosis and immunotherapy of pancreatic cancer.Front Immunol. 2025 Mar 7;16:1546332. doi: 10.3389/fimmu.2025.1546332. eCollection 2025. Front Immunol. 2025. PMID: 40124355 Free PMC article. Review.
Cited by
-
Development of liquid biopsy in detection and screening of pancreatic cancer.Front Oncol. 2024 Jun 3;14:1415260. doi: 10.3389/fonc.2024.1415260. eCollection 2024. Front Oncol. 2024. PMID: 38887233 Free PMC article. Review.
-
Illuminating diabetes via multi-omics: Unraveling disease mechanisms and advancing personalized therapy.World J Diabetes. 2025 Jul 15;16(7):106218. doi: 10.4239/wjd.v16.i7.106218. World J Diabetes. 2025. PMID: 40697608 Free PMC article. Review.
-
Blood-based biomarkers in pancreatic ductal adenocarcinoma: developments over the last decade and what holds for the future- a review.Front Oncol. 2025 Apr 22;15:1555963. doi: 10.3389/fonc.2025.1555963. eCollection 2025. Front Oncol. 2025. PMID: 40330826 Free PMC article. Review.
-
Biomarkers for Early Detection of Pancreatic Cancer.Visc Med. 2025 May 28. doi: 10.1159/000546584. Online ahead of print. Visc Med. 2025. PMID: 40612538 Free PMC article. Review.
-
Liquid Biopsy in Pancreatic Ductal Adenocarcinoma: A Review of Methods and Applications.Int J Mol Sci. 2024 Oct 13;25(20):11013. doi: 10.3390/ijms252011013. Int J Mol Sci. 2024. PMID: 39456796 Free PMC article. Review.
References
-
- Zeng H, Chen W, Zheng R, Zhang S, Ji JS, Zou X, et al. Changing cancer survival in China during 2003–15: a pooled analysis of 17 population‐based cancer registries. Lancet Glob Health. 2018;6:e555–e567. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

