Association between novel genetic variants of Notch signaling pathway genes and survival of hepatitis B virus-related hepatocellular carcinoma
- PMID: 38562021
- PMCID: PMC10985410
- DOI: 10.1002/cam4.7040
Association between novel genetic variants of Notch signaling pathway genes and survival of hepatitis B virus-related hepatocellular carcinoma
Abstract
Background: Although the Notch pathway plays an important role in formation and progression of hepatocellular carcinoma (HCC), few studies have reported the associations between functional genetic variants and the survival of hepatitis B virus (HBV)-related HCC.
Methods: In the present study, we performed multivariable Cox proportional hazard regression analysis to evaluate associations between 36,101 SNPs in 264 Notch pathway-related genes and overall survival (OS) of 866 patients with HBV-related HCC.
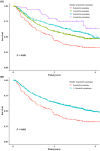
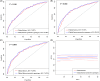
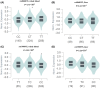
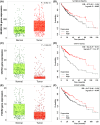
Results: It was found that three independent SNPs (NEURL1B rs4868192, CNTN1 rs444927 and FCER2 rs1990975) were significantly associated with the HBV-related HCC OS. The number of protective genotypes (NPGs) were significantly associated with better survival in a dose-response manner (ptrend <0.001). Compared with the model with sole clinical factors, the addition of protective genotypes to the predict models significantly increased the AUC, i.e., from 72.72% to 75.13% (p = 0.002) and from 72.04% to 74.76 (p = 0.004) for 3-year and 5-year OS, respectively. The expression quantitative trait loci (eQTL) analysis further revealed that the rs4868192 C allele was associated with lower mRNA expression levels of NEURL1B in the whole blood (p = 1.71 × 10-3), while the rs1990975 T allele was correlated with higher mRNA expression levels of FCER2 in the whole blood and normal liver tissues (p = 3.51 × 10-5 and 0.033, respectively).
Conclusions: Three potentially functional SNPs of NEURL1B, CNTN1 and FCER2 may serve as potential prognostic biomarkers for HBV-related HCC.
Keywords: Notch signaling pathway; hepatitis B virus; hepatocellular carcinoma; single nucleotide polymorphism; survival.
© 2024 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflict of interest.
Figures

Similar articles
-
Potentially functional genetic variants in RPS6KA4 and MAP2K5 in the MAPK signaling pathway predict HBV-related hepatocellular carcinoma survival.Mol Carcinog. 2023 Sep;62(9):1378-1387. doi: 10.1002/mc.23583. Epub 2023 Jun 6. Mol Carcinog. 2023. PMID: 37278562
-
Multi-omics integration analysis of the amino-acid metabolism-related genes identifies putatively causal variants of ACCS associated with hepatitis B virus-related hepatocellular carcinoma survival.BMC Cancer. 2025 Feb 18;25(1):284. doi: 10.1186/s12885-025-13604-3. BMC Cancer. 2025. PMID: 39966747 Free PMC article.
-
Prognostic value of Notch receptors in postsurgical patients with hepatitis B virus-related hepatocellular carcinoma.Cancer Med. 2017 Jul;6(7):1587-1600. doi: 10.1002/cam4.1077. Epub 2017 May 31. Cancer Med. 2017. PMID: 28568708 Free PMC article.
-
Blood-Based Biomarkers in Hepatitis B Virus-Related Hepatocellular Carcinoma, Including the Viral Genome and Glycosylated Proteins.Int J Mol Sci. 2021 Oct 13;22(20):11051. doi: 10.3390/ijms222011051. Int J Mol Sci. 2021. PMID: 34681709 Free PMC article. Review.
-
Single nucleotide polymorphisms associated with hepatocellular carcinoma in patients with chronic hepatitis B virus infection.Intervirology. 2005;48(1):10-5. doi: 10.1159/000082089. Intervirology. 2005. PMID: 15785084 Review.
References
-
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. - PubMed
-
- Sagnelli E, Macera M, Russo A, Coppola N, Sagnelli C. Epidemiological and etiological variations in hepatocellular carcinoma. Infection. 2020;48(1):7‐17. - PubMed
-
- Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet. 2022;400(10360):1345‐1362. - PubMed
-
- Yu Y, Mao L, Lu X, et al. Functional variant in 3'UTR of FAM13A is potentially associated with susceptibility and survival of lung squamous carcinoma. DNA Cell Biol. 2019;38(11):1269‐1277. - PubMed
MeSH terms
Substances
Grants and funding
- GKE-KF202102/Key Laboratory of Early Prevention and Treatment for Regional High Frequency Tumor, Ministry of Education
- GKE-ZZ202104/Key Laboratory of Early Prevention and Treatment for Regional High Frequency Tumor, Ministry of Education
- GKE-ZZ202118/Key Laboratory of Early Prevention and Treatment for Regional High Frequency Tumor, Ministry of Education
- YQJ2022-5/Youth Program of Scientific Research Foundation of Guangxi Medical University Cancer Hospital
- S2021022/Guangxi Medical and health appropriate Technology opening and popularization and application project
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

