Comparative transcriptomics and proteomics analysis of the symbiotic germination of Paphiopedilum barbigerum with Epulorhiza sp. FQXY019
- PMID: 38562471
- PMCID: PMC10982344
- DOI: 10.3389/fmicb.2024.1358137
Comparative transcriptomics and proteomics analysis of the symbiotic germination of Paphiopedilum barbigerum with Epulorhiza sp. FQXY019
Abstract
Introduction: Paphiopedilum barbigerum is currently the rarest and most endangered species of orchids in China and has significant ornamental value. The mature seeds of P. barbigerum are difficult to germinate owing to the absence of an endosperm and are highly dependent on mycorrhizal fungi for germination and subsequent development. However, little is known about the regulation mechanisms of symbiosis and symbiotic germination of P. barbigerum seeds.
Methods: Herein, transcriptomics and proteomics were used to explore the changes in the P. barbigerum seeds after inoculation with (FQXY019 treatment group) or without (control group) Epulorhiza sp. FQXY019 at 90 days after germination.
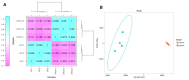
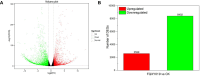
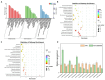
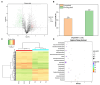
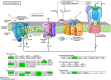
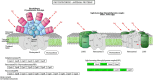
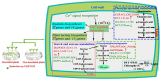
Results: Transcriptome sequencing revealed that a total of 10,961 differentially expressed genes (DEGs; 2,599 upregulated and 8,402 downregulated) were identified in the control and FQXY019 treatment groups. These DEGs were mainly involved in carbohydrate, fatty acid, and amino acid metabolism. Furthermore, the expression levels of candidate DEGs related to nodulin, Ca2+ signaling, and plant lectins were significantly affected in P. barbigerum in the FQXY019 treatment groups. Subsequently, tandem mass tag-based quantitative proteomics was performed to recognize the differentially expressed proteins (DEPs), and a total of 537 DEPs (220 upregulated and 317 downregulated) were identified that were enriched in processes including photosynthesis, photosynthesis-antenna proteins, and fatty acid biosynthesis and metabolism.
Discussion: This study provides novel insight on the mechanisms underlying the in vitro seed germination and protocorm development of P. barbigerum by using a compatible fungal symbiont and will benefit the reintroduction and mycorrhizal symbiotic germination of endangered orchids.
Keywords: Paphiopedilum barbigerum; mycorrhizal fungi; proteome; symbiotic germination; transcriptome.
Copyright © 2024 Tian, Wang, Ding, Wang, Yang, Bai, Tan and Liao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Isolation and identification of beneficial orchid mycorrhizal fungi in Paphiopedilum barbigerum (Orchidaceae).Plant Signal Behav. 2022 Dec 31;17(1):2005882. doi: 10.1080/15592324.2021.2005882. Epub 2021 Dec 16. Plant Signal Behav. 2022. PMID: 34913407 Free PMC article.
-
Symbiotic seed germination and seedling growth of mycorrhizal fungi in Paphiopedilum hirsutissimun (Lindl.Ex Hook.) Stein from China.Plant Signal Behav. 2023 Dec 31;18(1):2293405. doi: 10.1080/15592324.2023.2293405. Epub 2023 Dec 17. Plant Signal Behav. 2023. PMID: 38104263 Free PMC article.
-
Functional Insights into the Roles of Hormones in the Dendrobium officinale-Tulasnella sp. Germinated Seed Symbiotic Association.Int J Mol Sci. 2018 Nov 6;19(11):3484. doi: 10.3390/ijms19113484. Int J Mol Sci. 2018. PMID: 30404159 Free PMC article.
-
Orchid Reintroduction Based on Seed Germination-Promoting Mycorrhizal Fungi Derived From Protocorms or Seedlings.Front Plant Sci. 2021 Jun 30;12:701152. doi: 10.3389/fpls.2021.701152. eCollection 2021. Front Plant Sci. 2021. PMID: 34276753 Free PMC article. Review.
-
In Vitro Symbiotic Germination: A Revitalized Heuristic Approach for Orchid Species Conservation.Plants (Basel). 2020 Dec 9;9(12):1742. doi: 10.3390/plants9121742. Plants (Basel). 2020. PMID: 33317200 Free PMC article. Review.
Cited by
-
Unveiling the rhizosphere microbiome of Dendrobium: mechanisms, microbial interactions, and implications for sustainable agriculture.Front Microbiol. 2025 Jan 29;16:1531900. doi: 10.3389/fmicb.2025.1531900. eCollection 2025. Front Microbiol. 2025. PMID: 39944638 Free PMC article. Review.
References
-
- Andrade L. M., Peixoto-Junior R. F., Ribeiro R. V., Nóbile P. M., Brito M. S., Marchiori P. E. R. (2019). Biomass accumulation and cell wall structure of rice plants overexpressing a dirigent-jacalin of sugarcane (ShDJ) under varying conditions of water availability. Front. Plant Sci. 10:65. doi: 10.3389/fpls.2019.00065, PMID: - DOI - PMC - PubMed
-
- Balilashaki K., Zakizadeh H., Olfati J. A., Vahedi M., Kumar A., Indracanti M. (2019). Recent advances in Phalaenopsis orchid improvement using omics approaches. Plant Tissue Cult. Biotechnol. 29, 133–149. doi: 10.3329/ptcb.v29i1.41986 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

