Pathogenomes and virulence profiles of representative big six non-O157 serogroup Shiga toxin-producing Escherichia coli
- PMID: 38562479
- PMCID: PMC10982417
- DOI: 10.3389/fmicb.2024.1364026
Pathogenomes and virulence profiles of representative big six non-O157 serogroup Shiga toxin-producing Escherichia coli
Abstract
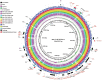
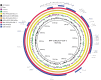
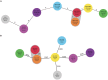
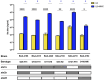
Shiga toxin (Stx)-producing Escherichia coli (STEC) of non-O157:H7 serotypes are responsible for global and widespread human food-borne disease. Among these serogroups, O26, O45, O103, O111, O121, and O145 account for the majority of clinical infections and are colloquially referred to as the "Big Six." The "Big Six" strain panel we sequenced and analyzed in this study are reference type cultures comprised of six strains representing each of the non-O157 STEC serogroups curated and distributed by the American Type Culture Collection (ATCC) as a resource to the research community under panel number ATCC MP-9. The application of long- and short-read hybrid sequencing yielded closed chromosomes and a total of 14 plasmids of diverse functions. Through high-resolution comparative phylogenomics, we cataloged the shared and strain-specific virulence and resistance gene content and established the close relationship of serogroup O26 and O103 strains featuring flagellar H-type 11. Virulence phenotyping revealed statistically significant differences in the Stx-production capabilities that we found to be correlated to the strain's individual stx-status. Among the carried Stx1a, Stx2a, and Stx2d phages, the Stx2a phage is by far the most responsive upon RecA-mediated phage mobilization, and in consequence, stx2a + isolates produced the highest-level of toxin in this panel. The availability of high-quality closed genomes for this "Big Six" reference set, including carried plasmids, along with the recorded genomic virulence profiles and Stx-production phenotypes will provide a valuable foundation to further explore the plasticity in evolutionary trajectories in these emerging non-O157 STEC lineages, which are major culprits of human food-borne disease.
Keywords: Shiga toxin (Stx)-producing Escherichia coli (STEC); non-O157 big six serogroups; phylogenomics; virulence phenotyping; whole genome sequencing and typing (WGST).
Copyright © 2024 Kalalah, Koenig, Bono, Bosilevac and Eppinger.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Shiga Toxin-Producing Escherichia coli (STEC) in Developing Countries: A 10-Year Review with Global Perspective.Microorganisms. 2025 Jun 30;13(7):1529. doi: 10.3390/microorganisms13071529. Microorganisms. 2025. PMID: 40732038 Free PMC article. Review.
-
Pathogenomes of Shiga Toxin Positive and Negative Escherichia coli O157:H7 Strains TT12A and TT12B: Comprehensive Phylogenomic Analysis Using Closed Genomes.Microorganisms. 2024 Mar 29;12(4):699. doi: 10.3390/microorganisms12040699. Microorganisms. 2024. PMID: 38674643 Free PMC article.
-
Genomic diversity of antimicrobial-resistant and Shiga toxin gene-harboring non-O157 Escherichia coli from dairy calves.J Glob Antimicrob Resist. 2023 Jun;33:164-170. doi: 10.1016/j.jgar.2023.02.022. Epub 2023 Mar 9. J Glob Antimicrob Resist. 2023. PMID: 36898633
-
A systematic review and meta-analysis of published literature on prevalence of non-O157 Shiga toxin-producing Escherichia coli serogroups (O26, O45, O103, O111, O121, and O145) and virulence genes in feces, hides, and carcasses of pre- and peri-harvest cattle worldwide.Anim Health Res Rev. 2022 Jun;23(1):1-24. doi: 10.1017/S1466252321000153. Epub 2022 Jun 9. Anim Health Res Rev. 2022. PMID: 35678500
-
Virulent shiga toxin-producing Escherichia coli (STEC) O157:H7 ST11 isolated from ground beef in Brazil.Braz J Microbiol. 2024 Dec;55(4):3513-3520. doi: 10.1007/s42770-024-01468-x. Epub 2024 Jul 31. Braz J Microbiol. 2024. PMID: 39083224 Free PMC article.
Cited by
-
Shiga toxin-producing Escherichia coli plasmid diversity reveals virulence potential and control opportunities in animal hosts.Sci Rep. 2025 Jul 22;15(1):26562. doi: 10.1038/s41598-025-11905-y. Sci Rep. 2025. PMID: 40696010 Free PMC article.
-
Shiga Toxin-Producing Escherichia coli (STEC) in Developing Countries: A 10-Year Review with Global Perspective.Microorganisms. 2025 Jun 30;13(7):1529. doi: 10.3390/microorganisms13071529. Microorganisms. 2025. PMID: 40732038 Free PMC article. Review.
-
Genotypic analysis of Shiga toxin-producing Escherichia coli clonal complex 17 in England and Wales, 2014-2022.J Med Microbiol. 2024 Nov;73(11):001928. doi: 10.1099/jmm.0.001928. J Med Microbiol. 2024. PMID: 39508726 Free PMC article.
-
An anti-Shiga toxin VHH nanobody multimer protects mice against fatal toxicosis when administered intramuscularly as repRNA.Infect Immun. 2024 Nov 12;92(11):e0023924. doi: 10.1128/iai.00239-24. Epub 2024 Oct 11. Infect Immun. 2024. PMID: 39392311 Free PMC article.
References
-
- AbuOun M., Jones H., Stubberfield E., Gilson D., Shaw L. P., Hubbard A. T. M., et al. . (2021). A genomic epidemiological study shows that prevalence of antimicrobial resistance in Enterobacterales is associated with the livestock host, as well as antimicrobial usage. Microb Genom 7:630. doi: 10.1099/mgen.0.000630 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

