Human protein-protein interaction networks: A topological comparison review
- PMID: 38562502
- PMCID: PMC10982977
- DOI: 10.1016/j.heliyon.2024.e27278
Human protein-protein interaction networks: A topological comparison review
Abstract
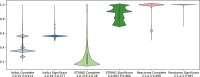
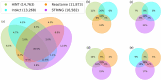
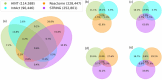
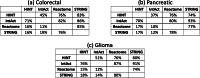
Protein-Protein Interaction Networks aim to model the interactome, providing a powerful tool for understanding the complex relationships governing cellular processes. These networks have numerous applications, including functional enrichment, discovering cancer driver genes, identifying drug targets, and more. Various databases make protein-protein networks available for many species, including Homo sapiens. This work topologically compares four Homo sapiens networks using a coarse-to-fine approach, comparing global characteristics, sub-network topology, specific nodes centrality, and interaction significance. Results show that the four human protein networks share many common protein-encoding genes and some global measures, but significantly differ in the interactions and neighbourhood. Small sub-networks from cancer pathways performed better than the whole networks, indicating an improved topological consistency in functional pathways. The centrality analysis shows that the same genes play different roles in different networks. We discuss how studies and analyses that rely on protein-protein networks for humans should consider their similarities and distinctions.
Keywords: Centrality measures; Complex systems; Network topology; PPIN; Protein–protein interaction networks.
© 2024 The Authors.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Rodrigo Henrique Ramos reports financial support and equipment, drugs, or supplies were provided by 10.13039/501100001807São Paulo Research Foundation (FAPESP). Rodrigo Henrique Ramos reports financial support and equipment, drugs, or supplies were provided by 10.13039/501100015897Center for Mathematical Sciences Applied to Industry (CeMEAI). Rodrigo Henrique Ramos reports financial support was provided by Brazilian National Research and Technology Council (10.13039/501100003593CNPq). Rodrigo Henrique Ramos reports financial support was provided by Brazilian Federal Foundation for Support and Evaluation of Graduate Education (10.13039/501100002322CAPES). If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures







Similar articles
-
A systematic survey of centrality measures for protein-protein interaction networks.BMC Syst Biol. 2018 Jul 31;12(1):80. doi: 10.1186/s12918-018-0598-2. BMC Syst Biol. 2018. PMID: 30064421 Free PMC article.
-
Consistency and differences between centrality measures across distinct classes of networks.PLoS One. 2019 Jul 26;14(7):e0220061. doi: 10.1371/journal.pone.0220061. eCollection 2019. PLoS One. 2019. PMID: 31348798 Free PMC article.
-
Examination of the relationship between essential genes in PPI network and hub proteins in reverse nearest neighbor topology.BMC Bioinformatics. 2010 Oct 12;11:505. doi: 10.1186/1471-2105-11-505. BMC Bioinformatics. 2010. PMID: 20939873 Free PMC article.
-
An integrative methodology based on protein-protein interaction networks for identification and functional annotation of disease-relevant genes applied to channelopathies.BMC Bioinformatics. 2019 Nov 12;20(1):565. doi: 10.1186/s12859-019-3162-1. BMC Bioinformatics. 2019. PMID: 31718537 Free PMC article.
-
Interaction networks: from protein functions to drug discovery. A review.Pathol Biol (Paris). 2009 Jun;57(4):324-33. doi: 10.1016/j.patbio.2008.10.004. Epub 2008 Dec 13. Pathol Biol (Paris). 2009. PMID: 19070972 Review.
Cited by
-
Longitudinal big biological data in the AI era.Mol Syst Biol. 2025 Aug 5. doi: 10.1038/s44320-025-00134-0. Online ahead of print. Mol Syst Biol. 2025. PMID: 40764831 Review.
-
The α/β3 complex of human voltage-gated sodium channel hNav1.7 to study mechanistic differences in presence and absence of auxiliary subunit β3.J Mol Model. 2025 May 21;31(6):168. doi: 10.1007/s00894-025-06378-9. J Mol Model. 2025. PMID: 40397258 Free PMC article.
-
State of the interactomes: an evaluation of molecular networks for generating biological insights.Mol Syst Biol. 2025 Jan;21(1):1-29. doi: 10.1038/s44320-024-00077-y. Epub 2024 Dec 9. Mol Syst Biol. 2025. PMID: 39653848 Free PMC article.
-
Identifying key genes in cancer networks using persistent homology.Sci Rep. 2025 Jan 22;15(1):2751. doi: 10.1038/s41598-025-87265-4. Sci Rep. 2025. PMID: 39838168 Free PMC article.
References
-
- Réka Albert, Barabási Albert-László. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002;74(1):47.
-
- Porras P. Network analysis of protein interaction data: an introduction. 2016. https://doi.org/10.6019/tol.networks_t.2016.00001.1 - DOI
-
- Koh Gavin C.K.W., et al. Analyzing protein–protein interaction networks. J. Proteome Res. 2012;11(4):2014–2031. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

