Oleuropein activates autophagy to circumvent anti-plasmodial defense
- PMID: 38562521
- PMCID: PMC10982566
- DOI: 10.1016/j.isci.2024.109463
Oleuropein activates autophagy to circumvent anti-plasmodial defense
Abstract
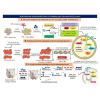
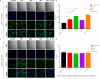
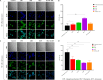
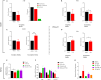
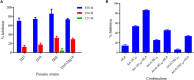
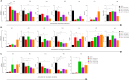
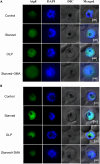
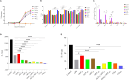
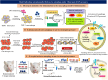
Antimalarial drug resistance and unavailability of effective vaccine warrant for newer drugs and drug targets. Hence, anti-inflammatory activity of phyto-compound (oleuropein; OLP) was determined in antigen (LPS)-stimulated human THP-1 macrophages (macrophage model of inflammation; MMI). Reduction in the inflammation was controlled by the PI3K-Akt1 signaling to establish the "immune-homeostasis." Also, OLP treatment influenced the cell death/autophagy axis leading to the modulated inflammation for extended cell survival. The findings with MII prompted us to detect the antimalarial activity of OLP in the wild type (3D7), D10-expressing GFP-Atg18 parasite, and chloroquine-resistant (Dd2) parasite. OLP did not show the parasite inhibition in the routine in vitro culture of P. falciparum whereas OLP increased the antimalarial activity of artesunate. The molecular docking of autophagy-related proteins, investigations with MMI, and parasite inhibition assays indicated that the host activated the autophagy to survive OLP pressure. The challenge model of P. berghei infection showed to induce autophagy for circumventing anti-plasmodial defenses.
Keywords: Drug delivery system; Health sciences.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Oleuropein mediated autophagy begets antimalarial drug resistance.Front Microbiol. 2024 Aug 20;15:1453998. doi: 10.3389/fmicb.2024.1453998. eCollection 2024. Front Microbiol. 2024. PMID: 39228384 Free PMC article.
-
Protocol for oleuropein-induced autophagy mediating drug tolerance in P. falciparum.STAR Protoc. 2024 Sep 20;5(3):103141. doi: 10.1016/j.xpro.2024.103141. Epub 2024 Jun 19. STAR Protoc. 2024. PMID: 38905105 Free PMC article.
-
Oleuropein: a viable therapeutic option for malaria and cancer.Drug Discov Today. 2025 Jan;30(1):104254. doi: 10.1016/j.drudis.2024.104254. Epub 2024 Nov 27. Drug Discov Today. 2025. PMID: 39608487 Review.
-
[In vitro susceptibility of P. falciparum isolates from Abidjan (Côte d'Ivoire) to quinine, artesunate and chloroquine].Sante. 2008 Jan-Mar;18(1):43-7. doi: 10.1684/san.2008.0103. Sante. 2008. PMID: 18684691 French.
-
[Combined antimalarial therapy using artemisinin].Parassitologia. 2004 Jun;46(1-2):85-7. Parassitologia. 2004. PMID: 15305693 Review. Italian.
Cited by
-
Biogenically synthesized green silver nanoparticles exhibit antimalarial activity.Discov Nano. 2024 Aug 31;19(1):136. doi: 10.1186/s11671-024-04098-2. Discov Nano. 2024. PMID: 39217276 Free PMC article.
-
From Soil to Brain: Olive Oil Attributes, Consumer Choices, Intermittent Fasting, and Their Impact on Health.Nutrients. 2025 Jun 1;17(11):1905. doi: 10.3390/nu17111905. Nutrients. 2025. PMID: 40507174 Free PMC article. Review.
-
Oleuropein mediated autophagy begets antimalarial drug resistance.Front Microbiol. 2024 Aug 20;15:1453998. doi: 10.3389/fmicb.2024.1453998. eCollection 2024. Front Microbiol. 2024. PMID: 39228384 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

