This is a preprint.
Asynchronous microexon splicing of LSD1 and PHF21A during neurodevelopment
- PMID: 38562691
- PMCID: PMC10983945
- DOI: 10.1101/2024.03.21.586181
Asynchronous microexon splicing of LSD1 and PHF21A during neurodevelopment
Update in
-
Neuronal splicing of the unmethylated histone H3K4 reader, PHF21A, prevents excessive synaptogenesis.J Biol Chem. 2024 Nov;300(11):107881. doi: 10.1016/j.jbc.2024.107881. Epub 2024 Oct 11. J Biol Chem. 2024. PMID: 39395799 Free PMC article.
Abstract
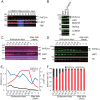
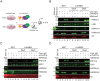
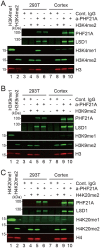
LSD1 histone H3K4 demethylase and its binding partner PHF21A, a reader protein for unmethylated H3K4, both undergo neuron-specific microexon splicing. The LSD1 neuronal microexon weakens H3K4 demethylation activity and can alter the substrate specificity to H3K9 or H4K20. Meanwhile, the PHF21A neuronal microexon interferes with nucleosome binding. However, the temporal expression patterns of LSD1 and PHF21A splicing isoforms during brain development remain unknown. In this work, we report that neuronal PHF21A isoform expression precedes neuronal LSD1 isoform expression during human neuron differentiation and mouse brain development. The asynchronous splicing events resulted in stepwise deactivation of the LSD1-PHF21A complex in reversing H3K4 methylation. We further show that the enzymatically inactive LSD1-PHF21A complex interacts with neuron-specific binding partners, including MYT1-family transcription factors and post-transcriptional mRNA processing proteins such as VIRMA. The interaction with the neuron-specific components, however, did not require the PHF21A microexon, indicating that the neuronal proteomic milieu, rather than the microexon-encoded PHF21A segment, is responsible for neuron-specific complex formation. These results indicate that the PHF21A microexon is dispensable for neuron-specific protein-protein interactions, yet the enzymatically inactive LSD1-PHF21A complex might have unique gene-regulatory roles in neurons.
Keywords: LSD1; PHF21A; histone demethylase; histone methylation; microexon splicing.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Najmabadi H., Hu H., Garshasbi M., Zemojtel T., Abedini S. S., Chen W., Hosseini M., Behjati F., Haas S., Jamali P., Zecha A., Mohseni M., Püttmann L., Vahid L. N., Jensen C., Moheb L. A., Bienek M., Larti F., Mueller I., Weissmann R., Darvish H., Wrogemann K., Hadavi V., Lipkowitz B., Esmaeeli-Nieh S., Wieczorek D., Kariminejad R., Firouzabadi S. G., Cohen M., Fattahi Z., Rost I., Mojahedi F., Hertzberg C., Dehghan A., Rajab A., Banavandi M. J. S., Hoffer J., Falah M., Musante L., Kalscheuer V., Ullmann R., Kuss A. W., Tzschach A., Kahrizi K., and Ropers H. H. (2011) Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 478, 57–63 - PubMed
-
- McCarthy S. E., Gillis J., Kramer M., Lihm J., Yoon S., Berstein Y., Mistry M., Pavlidis P., Solomon R., Ghiban E., Antoniou E., Kelleher E., O’Brien C., Donohoe G., Gill M., Morris D. W., McCombie W. R., and Corvin A. (2014) De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 19, 652–658 - PMC - PubMed
-
- De Rubeis S., He X., Goldberg A. P., Poultney C. S., Samocha K., Ercument Cicek A., Kou Y., Liu L., Fromer M., Walker S., Singh T., Klei L., Kosmicki J., Fu S.-C., Aleksic B., Biscaldi M., Bolton P. F., Brownfeld J. M., Cai J., Campbell N. G., Carracedo A., Chahrour M. H., Chiocchetti A. G., Coon H., Crawford E. L., Crooks L., Curran S. R., Dawson G., Duketis E., Fernandez B. A., Gallagher L., Geller E., Guter S. J., Sean Hill R., Ionita-Laza I., Jimenez Gonzalez P., Kilpinen H., Klauck S. M., Kolevzon A., Lee I., Lei J., Lehtimäki T., Lin C.-F., Ma’ayan A., Marshall C. R., McInnes A. L., Neale B., Owen M. J., Ozaki N., Parellada M., Parr J. R., Purcell S., Puura K., Rajagopalan D., Rehnström K., Reichenberg A., Sabo A., Sachse M., Sanders S. J., Schafer C., Schulte-Rüther M., Skuse D., Stevens C., Szatmari P., Tammimies K., Valladares O., Voran A., Wang L.-S., Weiss L. A., Jeremy Willsey A., Yu T. W., Yuen R. K. C., Cook E. H., Freitag C. M., Gill M., Hultman C. M., Lehner T., Palotie A., Schellenberg G. D., Sklar P., State M. W., Sutcliffe J. S., Walsh C. A., Scherer S. W., Zwick M. E., Barrett J. C., Cutler D. J., Roeder K., Devlin B., Daly M. J., and Buxbaum J. D. (2014) Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 515, 209–215 - PMC - PubMed
-
- Lister R., Mukamel E. A., Nery J. R., Urich M., Puddifoot C. A., Johnson N. D., Lucero J., Huang Y., Dwork A. J., Schultz M. D., Yu M., Tonti-Filippini J., Heyn H., Hu S., Wu J. C., Rao A., Esteller M., He C., Haghighi F. G., Sejnowski T. J., Behrens M. M., and Ecker J. R. (2013) Global Epigenomic Reconfiguration During Mammalian Brain Development. Science. 341, 1237905. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
