This is a preprint.
A Developmental Mechanism to Regulate Alternative Polyadenylation in an Adult Stem Cell Lineage
- PMID: 38562704
- PMCID: PMC10983978
- DOI: 10.1101/2024.03.18.585561
A Developmental Mechanism to Regulate Alternative Polyadenylation in an Adult Stem Cell Lineage
Update in
-
A developmental mechanism to regulate alternative polyadenylation in an adult stem cell lineage.Genes Dev. 2024 Aug 20;38(13-14):655-674. doi: 10.1101/gad.351649.124. Genes Dev. 2024. PMID: 39111825 Free PMC article.
Abstract
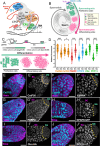
Alternative Cleavage and Polyadenylation (APA) often results in production of mRNA isoforms with either longer or shorter 3'UTRs from the same genetic locus, potentially impacting mRNA translation, localization and stability. Developmentally regulated APA can thus make major contributions to cell-type-specific gene expression programs as cells differentiate. During Drosophila spermatogenesis, approximately 500 genes undergo APA when proliferating spermatogonia differentiate into spermatocytes, producing transcripts with shortened 3' UTRs, leading to profound stage-specific changes in the proteins expressed. The molecular mechanisms that specify usage of upstream polyadenylation sites in spermatocytes are thus key to understanding the changes in cell state. Here, we show that upregulation of PCF11 and Cbc, the two components of Cleavage Factor II (CFII), orchestrates APA during Drosophila spermatogenesis. Knock down of PCF11 or cbc in spermatocytes caused dysregulation of APA, with many transcripts normally cleaved at a proximal site in spermatocytes now cleaved at their distal site, as in spermatogonia. Forced overexpression of CFII components in spermatogonia switched cleavage of some transcripts to the proximal site normally used in spermatocytes. Our findings reveal a developmental mechanism where changes in expression of specific cleavage factors can direct cell-type-specific APA at selected genes.
Keywords: Alternative Polyadenylation; Cellular Differentiation; Cleavage factor Complex II; Development; Drosophila; Spermatogenesis; mRNA processing.
Figures






Similar articles
-
A developmental mechanism to regulate alternative polyadenylation in an adult stem cell lineage.Genes Dev. 2024 Aug 20;38(13-14):655-674. doi: 10.1101/gad.351649.124. Genes Dev. 2024. PMID: 39111825 Free PMC article.
-
Developmentally regulated alternate 3' end cleavage of nascent transcripts controls dynamic changes in protein expression in an adult stem cell lineage.Genes Dev. 2022 Aug 1;36(15-16):916-935. doi: 10.1101/gad.349689.122. Epub 2022 Sep 29. Genes Dev. 2022. PMID: 36175033 Free PMC article.
-
The CFII components PCF11 and Cbc change subnuclear localization as cells differentiate in an adult stem cell lineage.MicroPubl Biol. 2025 Aug 15;2025:10.17912/micropub.biology.001718. doi: 10.17912/micropub.biology.001718. eCollection 2025. MicroPubl Biol. 2025. PMID: 40896257 Free PMC article.
-
Regulation and function of alternative polyadenylation in development and differentiation.RNA Biol. 2023 Jan;20(1):908-925. doi: 10.1080/15476286.2023.2275109. Epub 2023 Oct 31. RNA Biol. 2023. PMID: 37906624 Free PMC article. Review.
-
On the function and relevance of alternative 3'-UTRs in gene expression regulation.Wiley Interdiscip Rev RNA. 2021 Sep;12(5):e1653. doi: 10.1002/wrna.1653. Epub 2021 Apr 12. Wiley Interdiscip Rev RNA. 2021. PMID: 33843145 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
