This is a preprint.
Involvement of ArlI, ArlJ, and CirA in Archaeal Type-IV Pilin-Mediated Motility Regulation
- PMID: 38562816
- PMCID: PMC10983859
- DOI: 10.1101/2024.03.04.583388
Involvement of ArlI, ArlJ, and CirA in Archaeal Type-IV Pilin-Mediated Motility Regulation
Update in
-
Involvement of ArlI, ArlJ, and CirA in archaeal type IV pilin-mediated motility regulation.J Bacteriol. 2024 Jun 20;206(6):e0008924. doi: 10.1128/jb.00089-24. Epub 2024 May 31. J Bacteriol. 2024. PMID: 38819156 Free PMC article.
Abstract
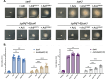
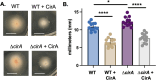
Many prokaryotes use swimming motility to move toward favorable conditions and escape adverse surroundings. Regulatory mechanisms governing bacterial flagella-driven motility are well-established, however, little is yet known about the regulation underlying swimming motility propelled by the archaeal cell surface structure, the archaella. Previous research showed that deletion of the adhesion pilins (PilA1-6), subunits of the type IV pili cell surface structure, renders the model archaeon Haloferax volcanii non-motile. In this study, we used EMS mutagenesis and a motility assay to identify motile suppressors of the ΔpilA[1-6] strain. Of the eight suppressors identified, six contain missense mutations in archaella biosynthesis genes, arlI and arlJ. Overexpression of these arlI and arlJ mutant constructs in the respective multi-deletion strains ΔpilA[1-6]ΔarlI and ΔpilA[1-6]ΔarlJ confirmed their role in suppressing the ΔpilA[1-6] motility defect. Additionally, three suppressors harbor co-occurring disruptive missense and nonsense mutations in cirA, a gene encoding a proposed regulatory protein. A deletion of cirA resulted in hypermotility, while cirA overexpression in wild-type cells led to decreased motility. Moreover, qRT-PCR analysis revealed that in wild-type cells, higher expression levels of arlI, arlJ, and the archaellin gene arlA1 were observed in motile early-log phase rod-shaped cells compared to non-motile mid-log phase disk-shaped cells. Conversely, ΔcirA cells, which form rods during both early and mid-log phases, exhibited similar expression levels of arl genes in both growth phases. Our findings contribute to a deeper understanding of the mechanisms governing archaeal motility, highlighting the involvement of ArlI, ArlJ, and CirA in pilin-mediated motility regulation.
Figures




Similar articles
-
Involvement of ArlI, ArlJ, and CirA in archaeal type IV pilin-mediated motility regulation.J Bacteriol. 2024 Jun 20;206(6):e0008924. doi: 10.1128/jb.00089-24. Epub 2024 May 31. J Bacteriol. 2024. PMID: 38819156 Free PMC article.
-
Identification of Haloferax volcanii Pilin N-Glycans with Diverse Roles in Pilus Biosynthesis, Adhesion, and Microcolony Formation.J Biol Chem. 2016 May 13;291(20):10602-14. doi: 10.1074/jbc.M115.693556. Epub 2016 Mar 10. J Biol Chem. 2016. PMID: 26966177 Free PMC article.
-
A conserved type IV pilin signal peptide H-domain is critical for the post-translational regulation of flagella-dependent motility.Mol Microbiol. 2014 Aug;93(3):494-504. doi: 10.1111/mmi.12673. Epub 2014 Jul 9. Mol Microbiol. 2014. PMID: 24945931
-
Diversity, assembly and regulation of archaeal type IV pili-like and non-type-IV pili-like surface structures.Res Microbiol. 2012 Nov-Dec;163(9-10):630-44. doi: 10.1016/j.resmic.2012.10.024. Epub 2012 Nov 9. Res Microbiol. 2012. PMID: 23146836 Review.
-
Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications.J Mol Microbiol Biotechnol. 2006;11(3-5):167-91. doi: 10.1159/000094053. J Mol Microbiol Biotechnol. 2006. PMID: 16983194 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
