This is a preprint.
A modular platform to generate functional sympathetic neuron-innervated heart assembloids
- PMID: 38562819
- PMCID: PMC10984094
- DOI: 10.21203/rs.3.rs-3894397/v1
A modular platform to generate functional sympathetic neuron-innervated heart assembloids
Abstract
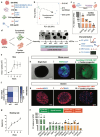
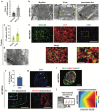
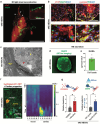
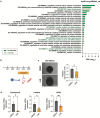
The technology of human pluripotent stem cell (hPSC)-based 3D organoid/assembloid cultures has become a powerful tool for the study of human embryonic development, disease modeling and drug discovery in recent years. The autonomic sympathetic nervous system innervates and regulates almost all organs in the body, including the heart. Yet, most reported organoids to date are not innervated, thus lacking proper neural regulation, and hindering reciprocal tissue maturation. Here, we developed a simple and versatile sympathetic neuron (symN)-innervated cardiac assembloid without the need for bioengineering. Our human sympathetic cardiac assembloids (hSCAs) showed mature muscle structures, atrial to ventricular patterning, and spontaneous beating. hSCA-innervating symNs displayed neurotransmitter synthesis and functional regulation of the cardiac beating rate, which could be manipulated pharmacologically or optogenetically. We modeled symN-mediated cardiac development and myocardial infarction. This hSCAs provides a tool for future neurocardiotoxicity screening approaches and is highly versatile and modular, where the types of neuron (symN or parasympathetic or sensory neuron) and organoid (heart, lung, kidney) to be innervated may be interchanged.
Conflict of interest statement
Confiict of interest We declare that the research was conducted without commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
O-GlcNAcylation is crucial for sympathetic neuron development, maintenance, functionality and contributes to peripheral neuropathy.Front Neurosci. 2023 May 5;17:1137847. doi: 10.3389/fnins.2023.1137847. eCollection 2023. Front Neurosci. 2023. PMID: 37229433 Free PMC article.
-
Engineering Human Brain Assembloids by Microfluidics.Adv Mater. 2023 Apr;35(14):e2210083. doi: 10.1002/adma.202210083. Epub 2023 Mar 2. Adv Mater. 2023. PMID: 36634089
-
Advances in Central Nervous System Organoids: A Focus on Organoid-Based Models for Motor Neuron Disease.Tissue Eng Part C Methods. 2021 Mar;27(3):213-224. doi: 10.1089/ten.TEC.2020.0337. Epub 2021 Mar 3. Tissue Eng Part C Methods. 2021. PMID: 33446055 Review.
-
Generation of human gastric assembloids from primary fetal organoids.Pediatr Surg Int. 2023 Nov 24;40(1):6. doi: 10.1007/s00383-023-05586-9. Pediatr Surg Int. 2023. PMID: 37999863 Free PMC article.
-
What Have Organoids and Assembloids Taught Us About the Pathophysiology of Neuropsychiatric Disorders?Biol Psychiatry. 2023 Apr 1;93(7):632-641. doi: 10.1016/j.biopsych.2022.11.017. Epub 2022 Dec 2. Biol Psychiatry. 2023. PMID: 36739210 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

