This is a preprint.
Germinal Center Dark Zone harbors ATR-dependent determinants of T-cell exclusion that are also identified in aggressive lymphoma
- PMID: 38562878
- PMCID: PMC10984086
- DOI: 10.21203/rs.3.rs-4093618/v1
Germinal Center Dark Zone harbors ATR-dependent determinants of T-cell exclusion that are also identified in aggressive lymphoma
Update in
-
Aggressive B cell lymphomas retain ATR-dependent determinants of T cell exclusion from the germinal center dark zone.J Clin Invest. 2025 Jul 17;135(18):e187371. doi: 10.1172/JCI187371. eCollection 2025 Sep 16. J Clin Invest. 2025. PMID: 40674145 Free PMC article.
Abstract
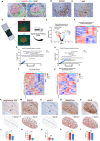
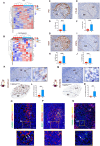
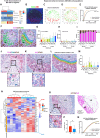
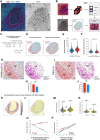
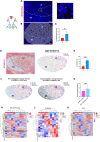
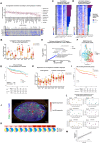
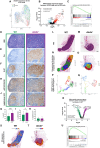
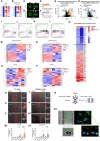
The germinal center (GC) dark zone (DZ) and light zone (LZ) regions spatially separate expansion and diversification from selection of antigen-specific B-cells to ensure antibody affinity maturation and B cell memory. The DZ and LZ differ significantly in their immune composition despite the lack of a physical barrier, yet the determinants of this polarization are poorly understood. This study provides novel insights into signals controlling asymmetric T-cell distribution between DZ and LZ regions. We identify spatially-resolved DNA damage response and chromatin compaction molecular features that underlie DZ T-cell exclusion. The DZ spatial transcriptional signature linked to T-cell immune evasion clustered aggressive Diffuse Large B-cell Lymphomas (DLBCL) for differential T cell infiltration. We reveal the dependence of the DZ transcriptional core signature on the ATR kinase and dissect its role in restraining inflammatory responses contributing to establishing an immune-repulsive imprint in DLBCL. These insights may guide ATR-focused treatment strategies bolstering immunotherapy in tumors marked by DZ transcriptional and chromatin-associated features.
Conflict of interest statement
CONFLICT OF INTERESTS ADJ has received consultancy fees from DKSH/Beigene, Roche, Gilead, Turbine Ltd, AstraZeneca, Antengene, Janssen, MSD and IQVIA; and research funding from Janssen and AstraZeneca.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

