This is a preprint.
Light-gated Integrator for Highlighting Kinase Activity in Living Cells
- PMID: 38562887
- PMCID: PMC10983958
- DOI: 10.1101/2024.03.18.585554
Light-gated Integrator for Highlighting Kinase Activity in Living Cells
Update in
-
Light-gated integrator for highlighting kinase activity in living cells.Nat Commun. 2024 Sep 6;15(1):7804. doi: 10.1038/s41467-024-51270-4. Nat Commun. 2024. PMID: 39242543 Free PMC article.
Abstract
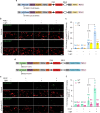
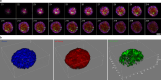
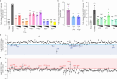
Protein kinases are key signaling nodes that regulate fundamental biological and disease processes. Illuminating kinase signaling from multiple angles can provide deeper insights into disease mechanisms and improve therapeutic targeting. While fluorescent biosensors are powerful tools for visualizing live-cell kinase activity dynamics in real time, new molecular tools are needed that enable recording of transient signaling activities for post hoc analysis and targeted manipulation. Here, we develop a light-gated kinase activity coupled transcriptional integrator (KINACT) that converts dynamic kinase signals into "permanent" fluorescent marks. KINACT enables robust monitoring of kinase activity across scales, accurately recording subcellular PKA activity, highlighting PKA signaling heterogeneity in 3D cultures, and identifying PKA activators and inhibitors in high-throughput screens. We further leverage the ability of KINACT to drive signaling effector expression to allow feedback manipulation of the balance of GαsR201C-induced PKA and ERK activation and dissect the mechanisms of oncogenic G protein signaling.
Figures


Similar articles
-
Light-gated integrator for highlighting kinase activity in living cells.Nat Commun. 2024 Sep 6;15(1):7804. doi: 10.1038/s41467-024-51270-4. Nat Commun. 2024. PMID: 39242543 Free PMC article.
-
Evaluating the predictive power of combined gene expression dynamics from single cells on antibiotic survival.mSystems. 2025 Jun 17;10(6):e0158824. doi: 10.1128/msystems.01588-24. Epub 2025 May 20. mSystems. 2025. PMID: 40391890 Free PMC article.
-
Prediction, screening and characterization of novel bioactive tetrapeptide matrikines for skin rejuvenation.Br J Dermatol. 2024 Jun 20;191(1):92-106. doi: 10.1093/bjd/ljae061. Br J Dermatol. 2024. PMID: 38375775
-
Defining disease severity in atopic dermatitis and psoriasis for the application to biomarker research: an interdisciplinary perspective.Br J Dermatol. 2024 Jun 20;191(1):14-23. doi: 10.1093/bjd/ljae080. Br J Dermatol. 2024. PMID: 38419411 Free PMC article. Review.
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
References
-
- Lee D., Hyun J.H., Jung K., Hannan P. & Kwon H.B. A calcium- and light-gated switch to induce gene expression in activated neurons. Nat. Biotechnol. 35, 858–863 (2017). - PubMed
-
- Lee D. et al. Temporally precise labeling and control of neuromodulatory circuits in the mammalian brain. Nat. Methods 14, 495–503 (2017). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
