This is a preprint.
Upstream open reading frames may contain hundreds of novel human exons
- PMID: 38562894
- PMCID: PMC10983949
- DOI: 10.1101/2024.03.22.586333
Upstream open reading frames may contain hundreds of novel human exons
Update in
-
Upstream open reading frames may contain hundreds of novel human exons.PLoS Comput Biol. 2024 Nov 20;20(11):e1012543. doi: 10.1371/journal.pcbi.1012543. eCollection 2024 Nov. PLoS Comput Biol. 2024. PMID: 39565752 Free PMC article.
Abstract
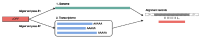
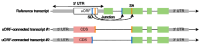
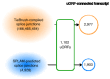
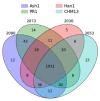
Several recent studies have presented evidence that the human gene catalogue should be expanded to include thousands of short open reading frames (ORFs) appearing upstream or downstream of existing protein-coding genes, each of which would comprise an additional bicistronic transcript in humans. Here we explore an alternative hypothesis that would explain the translational and evolutionary evidence for these upstream ORFs without the need to create novel genes or bicistronic transcripts. We examined 2,199 upstream ORFs that have been proposed as high-quality candidates for novel genes, to determine if they could instead represent protein-coding exons that can be added to existing genes. We checked for the conservation of these ORFs in four recently sequenced, high-quality human genomes, and found a large majority (87.8%) to be conserved in all four as expected. We then looked for splicing evidence that would connect each upstream ORF to the downstream protein-coding gene at the same locus, thus creating a novel splicing variant using the upstream ORF as its first exon. These protein coding exon candidates were further evaluated using protein structure predictions of the protein sequences that included the proposed new exons. We determined that 582 out of 2,199 upstream ORFs have strong evidence that they can form protein coding exons that are part of an existing gene, and that the resulting protein is predicted to have similar or better structural quality than the currently annotated isoform.
Figures
Similar articles
-
Upstream open reading frames may contain hundreds of novel human exons.PLoS Comput Biol. 2024 Nov 20;20(11):e1012543. doi: 10.1371/journal.pcbi.1012543. eCollection 2024 Nov. PLoS Comput Biol. 2024. PMID: 39565752 Free PMC article.
-
[Analysis, identification and correction of some errors of model refseqs appeared in NCBI Human Gene Database by in silico cloning and experimental verification of novel human genes].Yi Chuan Xue Bao. 2004 May;31(5):431-43. Yi Chuan Xue Bao. 2004. PMID: 15478601 Chinese.
-
Discovery of high-confidence human protein-coding genes and exons by whole-genome PhyloCSF helps elucidate 118 GWAS loci.Genome Res. 2019 Dec;29(12):2073-2087. doi: 10.1101/gr.246462.118. Epub 2019 Sep 19. Genome Res. 2019. PMID: 31537640 Free PMC article.
-
["Matreshka" Genes with Alternative Reading Frames].Genetika. 2016 Feb;52(2):146-63. Genetika. 2016. PMID: 27215029 Review. Russian.
-
The Protein-Coding Human Genome: Annotating High-Hanging Fruits.Bioessays. 2019 Nov;41(11):e1900066. doi: 10.1002/bies.201900066. Epub 2019 Sep 23. Bioessays. 2019. PMID: 31544971 Review.
References
-
- Calviello L., et al. (2016). “Detecting actively translated open reading frames in ribosome profiling data.” Nature methods 13(2): 165–170. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
