This is a preprint.
Speciation within the Anopheles gambiae complex: high-throughput whole genome sequencing reveals evidence of a putative new cryptic taxon in 'far-west' Africa
- PMID: 38562903
- PMCID: PMC10984024
- DOI: 10.21203/rs.3.rs-3914444/v1
Speciation within the Anopheles gambiae complex: high-throughput whole genome sequencing reveals evidence of a putative new cryptic taxon in 'far-west' Africa
Update in
-
Population genomic evidence of a putative 'far-west' African cryptic taxon in the Anopheles gambiae complex.Commun Biol. 2024 Sep 10;7(1):1115. doi: 10.1038/s42003-024-06809-y. Commun Biol. 2024. PMID: 39256556 Free PMC article.
Abstract
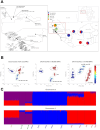
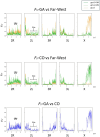
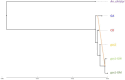
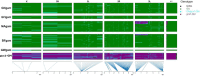
The two main Afrotropical malaria vectors - Anopheles coluzzii and An. gambiae - are genetically distinct and reproductively isolated across West Africa. However, populations at the western extreme of their range are assigned as "intermediate" between the two species by whole genome sequence (WGS) data, and as hybrid forms by conventional molecular diagnostics. By exploiting WGS data from 1,190 specimens collected across west Africa via the Anopheles gambiae 1000 Genomes network, we identify a novel putative taxon in the far-west (provisionally named Bissau molecular form), which did not arise by admixture but rather originated at the same time as the split between An. coluzzii and An. gambiae. Intriguingly, these populations lack insecticide resistance mechanisms commonly observed in the two main species. These findings lead to a change of perspective on malaria vector species in the far-west region with potential for epidemiological implications, and a new challenge for genetic-based mosquito control approaches.
Keywords: Anopheles; admixture; gene-flow; malaria vectors; reproductive barriers.
Conflict of interest statement
Additional Declarations: There is NO Competing Interest.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

