Prevalence of resistance-associated viral variants to the HIV-specific broadly neutralising antibody 10-1074 in a UK bNAb-naïve population
- PMID: 38562938
- PMCID: PMC10982389
- DOI: 10.3389/fimmu.2024.1352123
Prevalence of resistance-associated viral variants to the HIV-specific broadly neutralising antibody 10-1074 in a UK bNAb-naïve population
Abstract
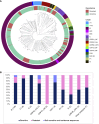
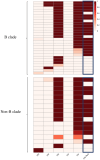
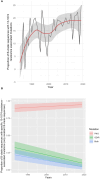
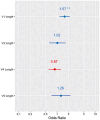
Broadly neutralising antibodies (bNAbs) targeting HIV show promise for both prevention of infection and treatment. Among these, 10-1074 has shown potential in neutralising a wide range of HIV strains. However, resistant viruses may limit the clinical efficacy of 10-1074. The prevalence of both de novo and emergent 10-1074 resistance will determine its use at a population level both to protect against HIV transmission and as an option for treatment. To help understand this further, we report the prevalence of pre-existing mutations associated with 10-1074 resistance in a bNAb-naive population of 157 individuals presenting to UK HIV centres with primary HIV infection, predominantly B clade, receiving antiretroviral treatment. Single genome analysis of HIV proviral envelope sequences showed that 29% of participants' viruses tested had at least one sequence with 10-1074 resistance-associated mutations. Mutations interfering with the glycan binding site at HIV Env position 332 accounted for 95% of all observed mutations. Subsequent analysis of a larger historic dataset of 2425 B-clade envelope sequences sampled from 1983 to 2019 revealed an increase of these mutations within the population over time. Clinical studies have shown that the presence of pre-existing bNAb mutations may predict diminished therapeutic effectiveness of 10-1074. Therefore, we emphasise the importance of screening for these mutations before initiating 10-1074 therapy, and to consider the implications of pre-existing resistance when designing prevention strategies.
Keywords: 10-1074; HIV - human immunodeficiency virus; broadly neutralising antibodies; primary HIV infection (PHI); resistance screening.
Copyright © 2024 Zacharopoulou, Lee, Oliveira, Thornhill, Robinson, Brown, Kinloch, Goulder, Fox, Fidler, Ansari and Frater.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Sensitivity to Broadly Neutralizing Antibodies of Recently Transmitted HIV-1 Clade CRF02_AG Viruses with a Focus on Evolution over Time.J Virol. 2019 Jan 4;93(2):e01492-18. doi: 10.1128/JVI.01492-18. Print 2019 Jan 15. J Virol. 2019. PMID: 30404804 Free PMC article.
-
Resistance mutations that distinguish HIV-1 envelopes with discordant VRC01 phenotypes from multi-lineage infections in the HVTN703/HPTN081 trial: implications for cross-resistance.J Virol. 2025 Feb 25;99(2):e0173024. doi: 10.1128/jvi.01730-24. Epub 2025 Jan 16. J Virol. 2025. PMID: 39817771 Free PMC article.
-
A Rare Mutation in an Infant-Derived HIV-1 Envelope Glycoprotein Alters Interprotomer Stability and Susceptibility to Broadly Neutralizing Antibodies Targeting the Trimer Apex.J Virol. 2020 Sep 15;94(19):e00814-20. doi: 10.1128/JVI.00814-20. Print 2020 Sep 15. J Virol. 2020. PMID: 32669335 Free PMC article.
-
Strategies for induction of HIV-1 envelope-reactive broadly neutralizing antibodies.J Int AIDS Soc. 2021 Nov;24 Suppl 7(Suppl 7):e25831. doi: 10.1002/jia2.25831. J Int AIDS Soc. 2021. PMID: 34806332 Free PMC article. Review.
-
Broadly neutralizing antibodies for the treatment and prevention of HIV infection.Curr Opin HIV AIDS. 2020 Jan;15(1):49-55. doi: 10.1097/COH.0000000000000600. Curr Opin HIV AIDS. 2020. PMID: 31764199 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

