STING-Pathway Inhibiting Nanoparticles (SPINs) as a Platform for Treatment of Inflammatory Diseases
- PMID: 38563162
- PMCID: PMC11337154
- DOI: 10.1021/acsabm.3c01305
STING-Pathway Inhibiting Nanoparticles (SPINs) as a Platform for Treatment of Inflammatory Diseases
Abstract
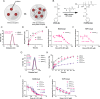
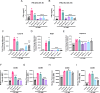
Aberrant activation of the cyclic GMP-AMP synthase (cGAS)/Stimulator of Interferon Genes (STING) pathway has been implicated in the development and progression of a myriad of inflammatory diseases including colitis, nonalcoholic steatohepatitis, amyotrophic lateral sclerosis (ALS), and age-related macular degeneration. Thus, STING pathway inhibitors could have therapeutic application in many of these inflammatory conditions. The cGAS inhibitor RU.521 and the STING inhibitor H-151 have shown promise as therapeutics in mouse models of colitis, ALS, and more. However, these agents require frequent high-dose intraperitoneal injections, which may limit translatability. Furthermore, long-term use of systemically administered cGAS/STING inhibitors may leave patients vulnerable to viral infections and cancer. Thus, localized or targeted inhibition of the cGAS/STING pathway may be an attractive, broadly applicable treatment for a variety of STING pathway-driven ailments. Here we describe STING-Pathway Inhibiting Nanoparticles (SPINS)-poly(lactic-co-glycolic acid) (PLGA) nanoparticles loaded with RU.521 and H-151-as a platform for enhanced and sustained inhibition of cGAS/STING signaling. We demonstrate that SPINs are equally or more effective at inhibiting type-I interferon responses induced by cytosolic DNA than free H-151 or RU.521. Additionally, we describe a SPIN formulation in which PLGA is coemulsified with poly(benzoyloxypropyl methacrylamide) (P(HPMA-Bz)), which significantly improves drug loading and allows for tunable release of H-151 over a period of days to over a week by varying P(HPMA-Bz) content. Finally, we find that all SPIN formulations were as potent or more potent in inhibiting cGAS/STING signaling in primary murine macrophages, resulting in decreased expression of inflammatory M1-like macrophage markers. Therefore, our study provides an in vitro proof-of-concept for nanoparticle delivery of STING pathway inhibitors and positions SPINs as a potential platform for slowing or reversing the onset or progression of cGAS/STING-driven inflammatory conditions.
Keywords: PLGA; STING; anti-inflammatory; cGAS; controlled release; inhibitor; nanoparticle.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
cGAS-STING and neurodegenerative diseases: A molecular crosstalk and therapeutic perspective.Int Immunopharmacol. 2025 Jun 26;159:114902. doi: 10.1016/j.intimp.2025.114902. Epub 2025 May 21. Int Immunopharmacol. 2025. PMID: 40403503 Review.
-
Oral Lipid-Based Nanomedicine for the Inhibition of the cGAS-STING Pathway in Inflammatory Bowel Disease Treatment.Mol Pharm. 2025 Apr 7;22(4):2108-2121. doi: 10.1021/acs.molpharmaceut.4c01297. Epub 2025 Mar 3. Mol Pharm. 2025. PMID: 40032274 Free PMC article.
-
Agonists and Inhibitors of the cGAS-STING Pathway.Molecules. 2024 Jun 30;29(13):3121. doi: 10.3390/molecules29133121. Molecules. 2024. PMID: 38999073 Free PMC article. Review.
-
Discovery of STING antagonists targeting cGAS-STING pathway to alleviate IMQ-induced psoriasis-like dermatitis.Eur J Pharm Sci. 2025 Jul 1;210:107091. doi: 10.1016/j.ejps.2025.107091. Epub 2025 Mar 31. Eur J Pharm Sci. 2025. PMID: 40174660
-
Morphine induces inflammatory responses via both TLR4 and cGAS-STING signaling pathways.Cytokine. 2024 Nov;183:156737. doi: 10.1016/j.cyto.2024.156737. Epub 2024 Aug 31. Cytokine. 2024. PMID: 39217915
Cited by
-
Inflammation-modulating polymeric nanoparticles: design strategies, mechanisms, and therapeutic applications.EBioMedicine. 2025 Aug;118:105837. doi: 10.1016/j.ebiom.2025.105837. Epub 2025 Jul 3. EBioMedicine. 2025. PMID: 40614330 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
