Effects of Klebsiella michiganensis LDS17 on Codonopsis pilosula growth, rhizosphere soil enzyme activities, and microflora, and genome-wide analysis of plant growth-promoting genes
- PMID: 38563743
- PMCID: PMC11064500
- DOI: 10.1128/spectrum.04056-23
Effects of Klebsiella michiganensis LDS17 on Codonopsis pilosula growth, rhizosphere soil enzyme activities, and microflora, and genome-wide analysis of plant growth-promoting genes
Abstract
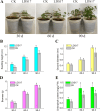
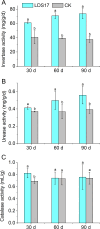
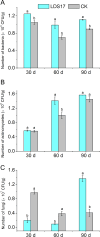
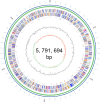
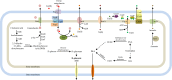
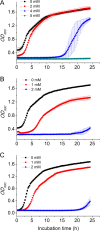
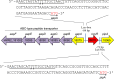
Codonopsis pilosula is a perennial herbaceous liana with medicinal value. It is critical to promote Codonopsis pilosula growth through effective and sustainable methods, and the use of plant growth-promoting bacteria (PGPB) is a promising candidate. In this study, we isolated a PGPB, Klebsiella michiganensis LDS17, that produced a highly active 1-aminocyclopropane-1-carboxylate deaminase from the Codonopsis pilosula rhizosphere. The strain exhibited multiple plant growth-promoting properties. The antagonistic activity of strain LDS17 against eight phytopathogenic fungi was investigated, and the results showed that strain LDS17 had obvious antagonistic effects on Rhizoctonia solani, Colletotrichum camelliae, Cytospora chrysosperma, and Phomopsis macrospore with growth inhibition rates of 54.22%, 49.41%, 48.89%, and 41.11%, respectively. Inoculation of strain LDS17 not only significantly increased the growth of Codonopsis pilosula seedlings but also increased the invertase and urease activities, the number of culturable bacteria, actinomycetes, and fungi, as well as the functional diversity of microbial communities in the rhizosphere soil of the seedlings. Heavy metal (HM) resistance tests showed that LDS17 is resistant to copper, zinc, and nickel. Whole-genome analysis of strain LDS17 revealed the genes involved in IAA production, siderophore synthesis, nitrogen fixation, P solubilization, and HM resistance. We further identified a gene (koyR) encoding a plant-responsive LuxR solo in the LDS17 genome. Klebsiella michiganensis LDS17 may therefore be useful in microbial fertilizers for Codonopsis pilosula. The identification of genes related to plant growth and HM resistance provides an important foundation for future analyses of the molecular mechanisms underlying the plant growth promotion and HM resistance of LDS17.
Importance: We comprehensively evaluated the plant growth-promoting characteristics and heavy metal (HM) resistance ability of the LDS17 strain, as well as the effects of strain LDS17 inoculation on the Codonopsis pilosula seedling growth and the soil qualities in the Codonopsis pilosula rhizosphere. We conducted whole-genome analysis and identified lots of genes and gene clusters contributing to plant-beneficial functions and HM resistance, which is critical for further elucidating the plant growth-promoting mechanism of strain LDS17 and expanding its application in the development of plant growth-promoting agents used in the environment under HM stress.
Keywords: ACC deaminase; Klebsiella michiganensis; heavy metal resistance; microbial functional diversity; plant growth-promoting bacteria; plant growth-promoting property; soil enzyme activity; whole-genome analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Bacillus licheniformisYB06: A Rhizosphere-Genome-Wide Analysis and Plant Growth-Promoting Analysis of a Plant Growth-Promoting Rhizobacterium Isolated from Codonopsis pilosula.Microorganisms. 2024 Sep 8;12(9):1861. doi: 10.3390/microorganisms12091861. Microorganisms. 2024. PMID: 39338535 Free PMC article.
-
[Effect of Continuous Cropping on the Physicochemical Properties, Microbial Activity, and Community Characteristics of the Rhizosphere Soil of Codonopsis pilosula].Huan Jing Ke Xue. 2023 Nov 8;44(11):6387-6398. doi: 10.13227/j.hjkx.202211100. Huan Jing Ke Xue. 2023. PMID: 37973120 Chinese.
-
Characterization of Cd-resistant Klebsiella michiganensis MCC3089 and its potential for rice seedling growth promotion under Cd stress.Microbiol Res. 2018 May;210:12-25. doi: 10.1016/j.micres.2018.03.003. Epub 2018 Mar 8. Microbiol Res. 2018. PMID: 29625654
-
The role of drought response genes and plant growth promoting bacteria on plant growth promotion under sustainable agriculture: A review.Microbiol Res. 2024 Sep;286:127827. doi: 10.1016/j.micres.2024.127827. Epub 2024 Jul 4. Microbiol Res. 2024. PMID: 39002396 Review.
-
Reducing Drought Stress in Plants by Encapsulating Plant Growth-Promoting Bacteria with Polysaccharides.Int J Mol Sci. 2021 Nov 30;22(23):12979. doi: 10.3390/ijms222312979. Int J Mol Sci. 2021. PMID: 34884785 Free PMC article. Review.
Cited by
-
Nodules-associated Klebsiella oxytoca complex: genomic insights into plant growth promotion and health risk assessment.BMC Microbiol. 2025 May 15;25(1):294. doi: 10.1186/s12866-025-04002-7. BMC Microbiol. 2025. PMID: 40375127 Free PMC article.
-
The effects of Rhodopseudomonas palustris on the improvement of agronomic traits and key enzyme-coding genes related to polysaccharide biosynthesis in Codonopsis pilosula.PLoS One. 2025 Jun 3;20(6):e0319989. doi: 10.1371/journal.pone.0319989. eCollection 2025. PLoS One. 2025. PMID: 40460104 Free PMC article.
References
-
- Bashan Y, Holguin G. 1998. Proposal for the division of plant growth-promoting rhizobacteria into two classifications: biocontrol-PGPB (plant growth-promoting bacteria) and PGPB. Soil Biology and Biochemistry 30:1225–1228. doi:10.1016/S0038-0717(97)00187-9 - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 32000069/MOST | National Natural Science Foundation of China (NSFC)
- 32071770/MOST | National Natural Science Foundation of China (NSFC)
- 20230044/Fund Program for the Scientific Activities of Selected Returned Overseas Professionals in Shanxi Province
- 2023-172/Research Project supported by Shanxi Scholarship Council of China
- Fund for Shanxi Province "1331 Projects Construction
LinkOut - more resources
Full Text Sources

