NF-κB subunits RelA and c-Rel selectively control CD4+ T cell function in multiple sclerosis and cancer
- PMID: 38563819
- PMCID: PMC10986815
- DOI: 10.1084/jem.20231348
NF-κB subunits RelA and c-Rel selectively control CD4+ T cell function in multiple sclerosis and cancer
Abstract
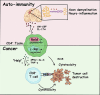
The outcome of cancer and autoimmunity is often dictated by the effector functions of CD4+ conventional T cells (Tconv). Although activation of the NF-κB signaling pathway has long been implicated in Tconv biology, the cell-autonomous roles of the separate NF-κB transcription-factor subunits are unknown. Here, we dissected the contributions of the canonical NF-κB subunits RelA and c-Rel to Tconv function. RelA, rather than c-Rel, regulated Tconv activation and cytokine production at steady-state and was required for polarization toward the TH17 lineage in vitro. Accordingly, RelA-deficient mice were fully protected against neuroinflammation in a model of multiple sclerosis due to defective transition to a pathogenic TH17 gene-expression program. Conversely, Tconv-restricted ablation of c-Rel impaired their function in the microenvironment of transplanted tumors, resulting in enhanced cancer burden. Moreover, Tconv required c-Rel for the response to PD-1-blockade therapy. Our data reveal distinct roles for canonical NF-κB subunits in different disease contexts, paving the way for subunit-targeted immunotherapies.
© 2024 Lalle et al.
Conflict of interest statement
Disclosures: P. Cassier reported “other” from Abbvie, Amgen, Alligator, Blueprint, Boehringer Ingelheim, Daiichi Sankyo, Astrazeneca, C4 Therapeutics, Exelixis, Molecular Partners, Pierre Fabre, Relay, Sotio, Tango, GSK, Merck Sharp Dohme, Lilly/Loxo, Novartis, Roche/Genentech, Taiho, Transgene, and Toray; personal fees from BMS, OSE, and Brenus; and non-financial support from Debio outside the submitted work. R. Liblau reported personal fees from Novartis, Biogen, Sanofi-genzyme, and Vida Venture; non-financial support from Novo Nordisk; and grants from Roche, Population Bio, and BMS outside the submitted work. No other disclosures were reported.
Figures










References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

