Individuals with JAK1 variants are affected by syndromic features encompassing autoimmunity, atopy, colitis, and dermatitis
- PMID: 38563820
- PMCID: PMC10986756
- DOI: 10.1084/jem.20232387
Individuals with JAK1 variants are affected by syndromic features encompassing autoimmunity, atopy, colitis, and dermatitis
Erratum in
-
Correction: Individuals with JAK1 variants are affected by syndromic features encompassing autoimmunity, atopy, colitis, and dermatitis.J Exp Med. 2024 Jun 3;221(6):e2023238704302024c. doi: 10.1084/jem.2023238704302024c. Epub 2024 May 6. J Exp Med. 2024. PMID: 38709237 Free PMC article. No abstract available.
Abstract
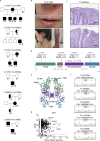
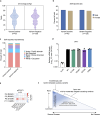
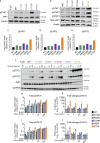
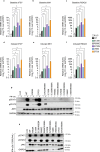
Inborn errors of immunity lead to autoimmunity, inflammation, allergy, infection, and/or malignancy. Disease-causing JAK1 gain-of-function (GoF) mutations are considered exceedingly rare and have been identified in only four families. Here, we use forward and reverse genetics to identify 59 individuals harboring one of four heterozygous JAK1 variants. In vitro and ex vivo analysis of these variants revealed hyperactive baseline and cytokine-induced STAT phosphorylation and interferon-stimulated gene (ISG) levels compared with wild-type JAK1. A systematic review of electronic health records from the BioME Biobank revealed increased likelihood of clinical presentation with autoimmunity, atopy, colitis, and/or dermatitis in JAK1 variant-positive individuals. Finally, treatment of one affected patient with severe atopic dermatitis using the JAK1/JAK2-selective inhibitor, baricitinib, resulted in clinically significant improvement. These findings suggest that individually rare JAK1 GoF variants may underlie an emerging syndrome with more common presentations of autoimmune and inflammatory disease (JAACD syndrome). More broadly, individuals who present with such conditions may benefit from genetic testing for the presence of JAK1 GoF variants.
© 2024 Bogunovic et al.
Conflict of interest statement
Disclosures: D. Bogunovic reported being owner and part owner of Lab11 Therapeutics. R.L. Fuleihan reported personal fees from Horizon, Takeda, Pharming, and Grifols outside the submitted work. I.K. Chinn reported other from Wolters Kluwer (UpToDate), Pharming, and Sumitomo Pharma America outside the submitted work. J.R. Lupski reported other from 23andMe and personal fees from Genome International, Inc, during the conduct of the study. N.S. Abul-Husn reported other from 23andMe and Allelica outside the submitted work. J.D. Milner reported personal fees from Blueprint Medicines outside the submitted work. No other disclosures were reported.
Figures



References
-
- Del Bel, K.L., Ragotte R.J., Saferali A., Lee S., Vercauteren S.M., Mostafavi S.A., Schreiber R.A., Prendiville J.S., Phang M.S., Halparin J., et al. . 2017. JAK1 gain-of-function causes an autosomal dominant immune dysregulatory and hypereosinophilic syndrome. J. Allergy Clin. Immunol. 139:2016–2020.e5. 10.1016/j.jaci.2016.12.957 - DOI - PubMed
-
- Fagerberg, L., Hallström B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., et al. . 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics. 13:397–406. 10.1074/mcp.M113.035600 - DOI - PMC - PubMed
-
- Geanon, D., Lee B., Gonzalez-Kozlova E., Kelly G., Handler D., Upadhyaya B., Leech J., De Real R.M., Herbinet M., Magen A., et al. . 2021. A streamlined whole blood CyTOF workflow defines a circulating immune cell signature of COVID-19. Cytometry. 99:446–461. 10.1002/cyto.a.24317 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

