NFĸB signaling drives myocardial injury via CCR2+ macrophages in a preclinical model of arrhythmogenic cardiomyopathy
- PMID: 38564300
- PMCID: PMC11093597
- DOI: 10.1172/JCI172014
NFĸB signaling drives myocardial injury via CCR2+ macrophages in a preclinical model of arrhythmogenic cardiomyopathy
Erratum in
-
NFĸB signaling drives myocardial injury via CCR2+ macrophages in a preclinical model of arrhythmogenic cardiomyopathy.J Clin Invest. 2024 Jul 1;134(13):e183441. doi: 10.1172/JCI183441. J Clin Invest. 2024. PMID: 38949031 Free PMC article. No abstract available.
Abstract
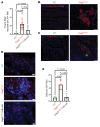
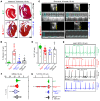
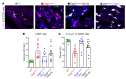
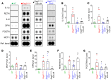
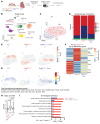
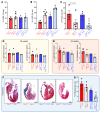
Nuclear factor κ-B (NFκB) is activated in iPSC-cardiac myocytes from patients with arrhythmogenic cardiomyopathy (ACM) under basal conditions, and inhibition of NFκB signaling prevents disease in Dsg2mut/mut mice, a robust mouse model of ACM. Here, we used genetic approaches and single-cell RNA-Seq to define the contributions of immune signaling in cardiac myocytes and macrophages in the natural progression of ACM using Dsg2mut/mut mice. We found that NFκB signaling in cardiac myocytes drives myocardial injury, contractile dysfunction, and arrhythmias in Dsg2mut/mut mice. NFκB signaling in cardiac myocytes mobilizes macrophages expressing C-C motif chemokine receptor-2 (CCR2+ cells) to affected areas within the heart, where they mediate myocardial injury and arrhythmias. Contractile dysfunction in Dsg2mut/mut mice is caused both by loss of heart muscle and negative inotropic effects of inflammation in viable muscle. Single nucleus RNA-Seq and cellular indexing of transcriptomes and epitomes (CITE-Seq) studies revealed marked proinflammatory changes in gene expression and the cellular landscape in hearts of Dsg2mut/mut mice involving cardiac myocytes, fibroblasts, and CCR2+ macrophages. Changes in gene expression in cardiac myocytes and fibroblasts in Dsg2mut/mut mice were dependent on CCR2+ macrophage recruitment to the heart. These results highlight complex mechanisms of immune injury and regulatory crosstalk between cardiac myocytes, inflammatory cells, and fibroblasts in the pathogenesis of ACM.
Keywords: Arrhythmias; Cardiology; Cardiovascular disease; Immunology; Innate immunity.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

