Filaggrin loss-of-function variants are associated with atopic dermatitis phenotypes in a diverse, early-life prospective cohort
- PMID: 38564302
- PMCID: PMC11141906
- DOI: 10.1172/jci.insight.178258
Filaggrin loss-of-function variants are associated with atopic dermatitis phenotypes in a diverse, early-life prospective cohort
Abstract
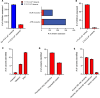
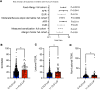
Loss-of-function (LoF) variants in the filaggrin (FLG) gene are the strongest known genetic risk factor for atopic dermatitis (AD), but the impact of these variants on AD outcomes is poorly understood. We comprehensively identified genetic variants through targeted region sequencing of FLG in children participating in the Mechanisms of Progression of Atopic Dermatitis to Asthma in Children cohort. Twenty FLG LoF variants were identified, including 1 novel variant and 9 variants not previously associated with AD. FLG LoF variants were found in the cohort. Among these children, the presence of 1 or more FLG LoF variants was associated with moderate/severe AD compared with those with mild AD. Children with FLG LoF variants had a higher SCORing for Atopic Dermatitis (SCORAD) and higher likelihood of food allergy within the first 2.5 years of life. LoF variants were associated with higher transepidermal water loss (TEWL) in both lesional and nonlesional skin. Collectively, our study identifies established and potentially novel AD-associated FLG LoF variants and associates FLG LoF variants with higher TEWL in lesional and nonlesional skin.
Keywords: Allergy; Dermatology; Genetics.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- U01 AI130830/AI/NIAID NIH HHS/United States
- R01 NS099068/NS/NINDS NIH HHS/United States
- R01 AI141569/AI/NIAID NIH HHS/United States
- R01 DK107502/DK/NIDDK NIH HHS/United States
- U01 AI150748/AI/NIAID NIH HHS/United States
- R01 HG010730/HG/NHGRI NIH HHS/United States
- R01 GM055479/GM/NIGMS NIH HHS/United States
- R01 AI024717/AI/NIAID NIH HHS/United States
- P30 AR070549/AR/NIAMS NIH HHS/United States
- R01 AI148276/AI/NIAID NIH HHS/United States
- U19 AI070235/AI/NIAID NIH HHS/United States
- P01 AI150585/AI/NIAID NIH HHS/United States
- R01 AR073228/AR/NIAMS NIH HHS/United States
- U01 HG011172/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

