IBDTransDB: a manually curated transcriptomic database for inflammatory bowel disease
- PMID: 38564306
- PMCID: PMC10986744
- DOI: 10.1093/database/baae026
IBDTransDB: a manually curated transcriptomic database for inflammatory bowel disease
Abstract
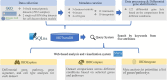
Inflammatory Bowel Disease (IBD) therapies are ineffective in at least 40% patients, and transcriptomic datasets have been widely used to reveal the pathogenesis and to identify the novel drug targets for these patients. Although public IBD transcriptomic datasets are available from many web-based tools/databases, due to the unstructured metadata and data description of these public datasets, most of these tools/databases do not allow querying datasets based on multiple keywords (e.g. colon and infliximab). Furthermore, few tools/databases can compare and integrate the datasets from the query results. To fill these gaps, we have developed IBDTransDB (https://abbviegrc.shinyapps.io/ibdtransdb/), a manually curated transcriptomic database for IBD. IBDTransDB includes a manually curated database with 34 transcriptomic datasets (2932 samples, 122 differential comparisons) and a query system supporting 35 keywords from 5 attributes (e.g. tissue and treatment). IBDTransDB also provides three modules for data analyses and integration. IBDExplore allows interactive visualization of differential gene list, pathway enrichment, gene signature and cell deconvolution analyses from a single dataset. IBDCompare supports comparisons of selected genes or pathways from multiple datasets across different conditions. IBDIntegrate performs meta-analysis to prioritize a list of genes/pathways based on user-selected datasets and conditions. Using two case studies related to infliximab treatment, we demonstrated that IBDTransDB provides a unique platform for biologists and clinicians to reveal IBD pathogenesis and identify the novel targets by integrating with other omics data. Database URL: https://abbviegrc.shinyapps.io/ibdtransdb/.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
JW, DC and PS are employees of AbbVie. SY is a contractor for AbbVie. VA was an employee of AbbVie at the time of the study. The design, study conduct and financial support for this research were provided by AbbVie. AbbVie participated in the interpretation of data, review and approval of the publication.
Figures


Similar articles
-
An Inflammatory Bowel Diseases Integrated Resources Portal (IBDIRP).Database (Oxford). 2024 Jan 16;2024:baad097. doi: 10.1093/database/baad097. Database (Oxford). 2024. PMID: 38227799 Free PMC article.
-
IBDsite: a Galaxy-interacting, integrative database for supporting inflammatory bowel disease high throughput data analysis.BMC Bioinformatics. 2012;13 Suppl 14(Suppl 14):S5. doi: 10.1186/1471-2105-13-S14-S5. Epub 2012 Sep 7. BMC Bioinformatics. 2012. PMID: 23095257 Free PMC article.
-
CTPAD: an interactive web application for comprehensive transcriptomic profiling in allergic diseases.J Transl Med. 2024 Oct 14;22(1):935. doi: 10.1186/s12967-024-05459-2. J Transl Med. 2024. PMID: 39402558 Free PMC article.
-
Potential shared pathogenic mechanisms between endometriosis and inflammatory bowel disease indicate a strong initial effect of immune factors.Front Immunol. 2024 Apr 3;15:1339647. doi: 10.3389/fimmu.2024.1339647. eCollection 2024. Front Immunol. 2024. PMID: 38660311 Free PMC article.
-
Expert recommendations to standardize transcriptomic analysis in inflammatory bowel disease clinical trials.J Crohns Colitis. 2025 May 8;19(5):jjaf068. doi: 10.1093/ecco-jcc/jjaf068. J Crohns Colitis. 2025. PMID: 40295219
References
-
- Torres J., Mehandru S., Colombel J.F. et al. (2017) Crohn’s disease. Lancet, 389, 1741–1755. - PubMed
-
- Wang J., Macoritto M., Guay H. et al. (2022) The clinical response of upadacitinib and risankizumab is associated with reduced inflammatory bowel disease anti-TNF-alpha inadequate response mechanisms. Inflamm. Bowel. Dis., 29, 771–782. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

