Nationwide genome surveillance of carbapenem-resistant Pseudomonas aeruginosa in Japan
- PMID: 38564665
- PMCID: PMC11064530
- DOI: 10.1128/aac.01669-23
Nationwide genome surveillance of carbapenem-resistant Pseudomonas aeruginosa in Japan
Abstract
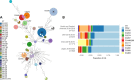
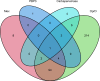
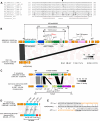
Japan is a country with an approximate 10% prevalence rate of carbapenem-resistant Pseudomonas aeruginosa (CRPA). Currently, a comprehensive overview of the genotype and phenotype patterns of CRPA in Japan is lacking. Herein, we conducted genome sequencing and quantitative antimicrobial susceptibility testing for 382 meropenem-resistant CRPA isolates that were collected from 78 hospitals across Japan from 2019 to 2020. CRPA exhibited susceptibility rates of 52.9%, 26.4%, and 88.0% against piperacillin-tazobactam, ciprofloxacin, and amikacin, respectively, whereas 27.7% of CRPA isolates was classified as difficult-to-treat resistance P. aeruginosa. Of the 148 sequence types detected, ST274 (9.7%) was predominant, followed by ST235 (7.6%). The proportion of urine isolates in ST235 was higher than that in other STs (P = 0.0056, χ2 test). Only 4.1% of CRPA isolates carried the carbapenemase genes: blaGES (2) and blaIMP (13). One ST235 isolate carried the novel blaIMP variant blaIMP-98 in the chromosome. Regarding chromosomal mutations, 87.1% of CRPA isolates possessed inactivating or other resistance mutations in oprD, and 28.8% showed mutations in the regulatory genes (mexR, nalC, and nalD) for the MexAB-OprM efflux pump. Additionally, 4.7% of CRPA isolates carried a resistance mutation in the PBP3-encoding gene ftsI. The findings from this study and other surveillance studies collectively demonstrate that CRPA exhibits marked genetic diversity and that its multidrug resistance in Japan is less prevailed than in other regions. This study contributes a valuable data set that addresses a gap in genotype/phenotype information regarding CRPA in the Asia-Pacific region, where the epidemiological background markedly differs between regions.
Keywords: DTR-P. aeruginosa; IMP; SE; carbapenemase; integron.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Senda K, Arakawa Y, Nakashima K, Ito H, Ichiyama S, Shimokata K, Kato N, Ohta M. 1996. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams, including carbapenems. Antimicrob Agents Chemother 40:349–353. doi: 10.1128/AAC.40.2.349 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

