A germline point mutation in the MYC-FBW7 phosphodegron initiates hematopoietic malignancies
- PMID: 38565249
- PMCID: PMC11065175
- DOI: 10.1101/gad.351292.123
A germline point mutation in the MYC-FBW7 phosphodegron initiates hematopoietic malignancies
Erratum in
-
Corrigendum: A germline point mutation in the MYC-FBW7 phosphodegron initiates hematopoietic malignancies.Genes Dev. 2025 Dec 1;39(23-24):1509. doi: 10.1101/gad.353345.125. Genes Dev. 2025. PMID: 41326179 Free PMC article. No abstract available.
Abstract
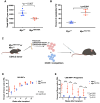
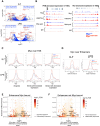
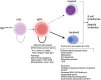
Oncogenic activation of MYC in cancers predominantly involves increased transcription rather than coding region mutations. However, MYC-dependent lymphomas frequently acquire point mutations in the MYC phosphodegron, including at threonine 58 (T58), where phosphorylation permits binding via the FBW7 ubiquitin ligase triggering MYC degradation. To understand how T58 phosphorylation functions in normal cell physiology, we introduced an alanine mutation at T58 (T58A) into the endogenous c-Myc locus in the mouse germline. While MYC-T58A mice develop normally, lymphomas and myeloid leukemias emerge in ∼60% of adult homozygous T58A mice. We found that primitive hematopoietic progenitor cells from MYC-T58A mice exhibit aberrant self-renewal normally associated with hematopoietic stem cells (HSCs) and up-regulate a subset of MYC target genes important in maintaining stem/progenitor cell balance. In lymphocytes, genomic occupancy by MYC-T58A was increased at all promoters compared with WT MYC, while genes differentially expressed in a T58A-dependent manner were significantly more proximal to MYC-bound enhancers. MYC-T58A lymphocyte progenitors exhibited metabolic alterations and decreased activation of inflammatory and apoptotic pathways. Our data demonstrate that a single point mutation stabilizing MYC is sufficient to skew target gene expression, producing a profound gain of function in multipotential hematopoietic progenitors associated with self-renewal and initiation of lymphomas and leukemias.
Keywords: FBW7; MYC; hematopoiesis; leukemia; lymphoma; progenitor cells; protein stability; self-renewal.
© 2024 Freie et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Update of
-
A Germline Point Mutation in the MYC-FBW7 Phosphodegron Initiates Hematopoietic Malignancies.bioRxiv [Preprint]. 2023 Oct 25:2023.10.23.563660. doi: 10.1101/2023.10.23.563660. bioRxiv. 2023. Update in: Genes Dev. 2024 Apr 17;38(5-6):253-272. doi: 10.1101/gad.351292.123. PMID: 37961183 Free PMC article. Updated. Preprint.
References
-
- Babcock JT, Nguyen HB, He Y, Hendricks JW, Wek RC, Quilliam LA. 2013. Mammalian target of rapamycin complex 1 (mTORC1) enhances bortezomib-induced death in tuberous sclerosis complex (TSC)-null cells by a c-MYC-dependent induction of the unfolded protein response. J Biol Chem 288: 15687–15698. 10.1074/jbc.M112.431056 - DOI - PMC - PubMed
-
- Belluschi S, Calderbank EF, Ciaurro V, Pijuan-Sala B, Santoro A, Mende N, Diamanti E, Sham KYC, Wang X, Lau WWY, et al. 2018. Myelo–lymphoid lineage restriction occurs in the human haematopoietic stem cell compartment before lymphoid-primed multipotent progenitors. Nat Commun 9: 4100. 10.1038/s41467-018-06442-4 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
