Hidden diversity and potential ecological function of phosphorus acquisition genes in widespread terrestrial bacteriophages
- PMID: 38565528
- PMCID: PMC10987575
- DOI: 10.1038/s41467-024-47214-7
Hidden diversity and potential ecological function of phosphorus acquisition genes in widespread terrestrial bacteriophages
Abstract
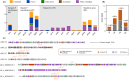
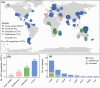
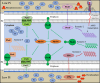
Phosphorus (P) limitation of ecosystem processes is widespread in terrestrial habitats. While a few auxiliary metabolic genes (AMGs) in bacteriophages from aquatic habitats are reported to have the potential to enhance P-acquisition ability of their hosts, little is known about the diversity and potential ecological function of P-acquisition genes encoded by terrestrial bacteriophages. Here, we analyze 333 soil metagenomes from five terrestrial habitat types across China and identify 75 viral operational taxonomic units (vOTUs) that encode 105 P-acquisition AMGs. These AMGs span 17 distinct functional genes involved in four primary processes of microbial P-acquisition. Among them, over 60% (11/17) have not been reported previously. We experimentally verify in-vitro enzymatic activities of two pyrophosphatases and one alkaline phosphatase encoded by P-acquisition vOTUs. Thirty-six percent of the 75 P-acquisition vOTUs are detectable in a published global topsoil metagenome dataset. Further analyses reveal that, under certain circumstances, the identified P-acquisition AMGs have a greater influence on soil P availability and are more dominant in soil metatranscriptomes than their corresponding bacterial genes. Overall, our results reinforce the necessity of incorporating viral contributions into biogeochemical P cycling.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Soil Viruses Are Underexplored Players in Ecosystem Carbon Processing.mSystems. 2018 Oct 2;3(5):e00076-18. doi: 10.1128/mSystems.00076-18. eCollection 2018 Sep-Oct. mSystems. 2018. PMID: 30320215 Free PMC article.
-
Viral community-wide auxiliary metabolic genes differ by lifestyles, habitats, and hosts.Microbiome. 2022 Nov 5;10(1):190. doi: 10.1186/s40168-022-01384-y. Microbiome. 2022. PMID: 36333738 Free PMC article.
-
Unraveling the habitat preferences, ecological drivers, potential hosts, and auxiliary metabolism of soil giant viruses across China.Microbiome. 2024 Jul 22;12(1):136. doi: 10.1186/s40168-024-01851-8. Microbiome. 2024. PMID: 39039586 Free PMC article.
-
Underexplored viral auxiliary metabolic genes in soil: Diversity and eco-evolutionary significance.Environ Microbiol. 2023 Apr;25(4):800-810. doi: 10.1111/1462-2920.16329. Epub 2023 Jan 5. Environ Microbiol. 2023. PMID: 36571495 Review.
-
Viral metabolic reprogramming in marine ecosystems.Curr Opin Microbiol. 2016 Jun;31:161-168. doi: 10.1016/j.mib.2016.04.002. Epub 2016 Apr 16. Curr Opin Microbiol. 2016. PMID: 27088500 Review.
Cited by
-
A call for caution in the biological interpretation of viral auxiliary metabolic genes.Nat Microbiol. 2025 Sep;10(9):2122-2129. doi: 10.1038/s41564-025-02095-4. Epub 2025 Aug 27. Nat Microbiol. 2025. PMID: 40866482 Review.
-
Rhizosphere-triggered viral lysogeny mediates microbial metabolic reprogramming to enhance arsenic oxidation.Nat Commun. 2025 Apr 30;16(1):4048. doi: 10.1038/s41467-025-58695-5. Nat Commun. 2025. PMID: 40307209 Free PMC article.
-
Viral Diversity in Polar Hydrocarbon-Contaminated Soils: A Transect Study from King George Island, Antarctica.Food Environ Virol. 2025 Aug 14;17(3):43. doi: 10.1007/s12560-025-09653-3. Food Environ Virol. 2025. PMID: 40810759
-
Unveiling the unknown viral world in groundwater.Nat Commun. 2024 Aug 8;15(1):6788. doi: 10.1038/s41467-024-51230-y. Nat Commun. 2024. PMID: 39117653 Free PMC article.
-
Revealing the Existence of Diverse Strategies for Phosphorus Solubilization and Acquisition in Plant-Growth Promoting Streptomyces misionensis SwB1.Microorganisms. 2025 Feb 9;13(2):378. doi: 10.3390/microorganisms13020378. Microorganisms. 2025. PMID: 40005744 Free PMC article.
References
-
- Morel C, Tunney P, Plenet D, Pellerin S. Transfer of phosphate ions between soil and solution: perspectives in soil testing. J. Environ. Qual. 2000;29:50–59. doi: 10.2134/jeq2000.00472425002900010007x. - DOI
-
- Cui YX, et al. Ecoenzymatic stoichiometry reveals widespread soil phosphorus limitation to microbial metabolism across Chinese forests. Commun. Earth Environ. 2022;3:184. doi: 10.1038/s43247-022-00523-5. - DOI
-
- Camenzind T, Hattenschwiler S, Treseder KK, Lehmann A, Rillig MC. Nutrient limitation of soil microbial processes in tropical forests. Ecol. Monogr. 2018;88:4–21. doi: 10.1002/ecm.1279. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

