The ER-SURF pathway uses ER-mitochondria contact sites for protein targeting to mitochondria
- PMID: 38565738
- PMCID: PMC11014988
- DOI: 10.1038/s44319-024-00113-w
The ER-SURF pathway uses ER-mitochondria contact sites for protein targeting to mitochondria
Abstract
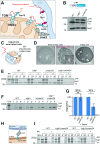
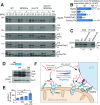
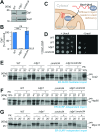
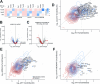
Most mitochondrial proteins are synthesized on cytosolic ribosomes and imported into mitochondria in a post-translational reaction. Mitochondrial precursor proteins which use the ER-SURF pathway employ the surface of the endoplasmic reticulum (ER) as an important sorting platform. How they reach the mitochondrial import machinery from the ER is not known. Here we show that mitochondrial contact sites play a crucial role in the ER-to-mitochondria transfer of precursor proteins. The ER mitochondria encounter structure (ERMES) and Tom70, together with Djp1 and Lam6, are part of two parallel and partially redundant ER-to-mitochondria delivery routes. When ER-to-mitochondria transfer is prevented by loss of these two contact sites, many precursors of mitochondrial inner membrane proteins are left stranded on the ER membrane, resulting in mitochondrial dysfunction. Our observations support an active role of the ER in mitochondrial protein biogenesis.
Keywords: Contact sites; ERMES; Endoplasmic reticulum; Mitochondria; Protein import.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







Similar articles
-
ER-SURF: Riding the Endoplasmic Reticulum Surface to Mitochondria.Int J Mol Sci. 2021 Sep 6;22(17):9655. doi: 10.3390/ijms22179655. Int J Mol Sci. 2021. PMID: 34502567 Free PMC article. Review.
-
Separating mitochondrial protein assembly and endoplasmic reticulum tethering by selective coupling of Mdm10.Nat Commun. 2016 Oct 10;7:13021. doi: 10.1038/ncomms13021. Nat Commun. 2016. PMID: 27721450 Free PMC article.
-
An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast.Science. 2018 Sep 14;361(6407):1118-1122. doi: 10.1126/science.aar8174. Science. 2018. PMID: 30213914
-
Connection of Protein Transport and Organelle Contact Sites in Mitochondria.J Mol Biol. 2017 Jul 7;429(14):2148-2160. doi: 10.1016/j.jmb.2017.05.023. Epub 2017 May 30. J Mol Biol. 2017. PMID: 28576471 Review.
-
ER exit sites (ERES) and ER-mitochondria encounter structures (ERMES) often localize proximally.FEBS Lett. 2023 Jan;597(2):320-336. doi: 10.1002/1873-3468.14497. Epub 2022 Oct 21. FEBS Lett. 2023. PMID: 36103135
Cited by
-
Cytoplasmic ribosomes on mitochondria alter the local membrane environment for protein import.J Cell Biol. 2025 Apr 7;224(4):e202407110. doi: 10.1083/jcb.202407110. Epub 2025 Mar 6. J Cell Biol. 2025. PMID: 40047641 Free PMC article.
-
J-domain proteins: From molecular mechanisms to diseases.Cell Stress Chaperones. 2024 Feb;29(1):21-33. doi: 10.1016/j.cstres.2023.12.002. Epub 2023 Dec 23. Cell Stress Chaperones. 2024. PMID: 38320449 Free PMC article.
-
Mitochondria, Peroxisomes and Beyond-How Dual Targeting Regulates Organelle Tethering.Contact (Thousand Oaks). 2024 Sep 28;7:25152564241264254. doi: 10.1177/25152564241264254. eCollection 2024 Jan-Dec. Contact (Thousand Oaks). 2024. PMID: 39364173 Free PMC article. Review.
-
Lipid scrambling is a general feature of protein insertases.bioRxiv [Preprint]. 2023 Sep 1:2023.09.01.555937. doi: 10.1101/2023.09.01.555937. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2024 Apr 23;121(17):e2319476121. doi: 10.1073/pnas.2319476121. PMID: 37693532 Free PMC article. Updated. Preprint.
-
Better Together: Interorganellar Communication in the Regulation of Proteostasis.Contact (Thousand Oaks). 2024 Oct 8;7:25152564241272245. doi: 10.1177/25152564241272245. eCollection 2024 Jan-Dec. Contact (Thousand Oaks). 2024. PMID: 39385949 Free PMC article. Review.
References
-
- Backes S, Bykov YS, Flohr T, Raschle M, Zhou J, Lenhard S, Kramer L, Muhlhaus T, Bibi C, Jann C, et al. The chaperone-binding activity of the mitochondrial surface receptor Tom70 protects the cytosol against mitoprotein-induced stress. Cell Rep. 2021;35:108936. doi: 10.1016/j.celrep.2021.108936. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 101052639/EC | ERC | HORIZON EUROPE European Research Council (ERC)
- HE2803/10-1/Deutsche Forschungsgemeinschaft (DFG)
- GRK2737/Deutsche Forschungsgemeinschaft (DFG)
- BioComp/Landesforschungsinitiative Rheinland-Pfalz
- 310030_185127/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (SNF)
LinkOut - more resources
Full Text Sources

