AI is a viable alternative to high throughput screening: a 318-target study
- PMID: 38565852
- PMCID: PMC10987645
- DOI: 10.1038/s41598-024-54655-z
AI is a viable alternative to high throughput screening: a 318-target study
Erratum in
-
Author Correction: AI is a viable alternative to high throughput screening: a 318-target study.Sci Rep. 2024 Sep 16;14(1):21579. doi: 10.1038/s41598-024-70321-w. Sci Rep. 2024. PMID: 39284864 Free PMC article. No abstract available.
Abstract
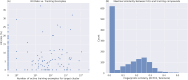
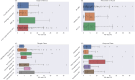
High throughput screening (HTS) is routinely used to identify bioactive small molecules. This requires physical compounds, which limits coverage of accessible chemical space. Computational approaches combined with vast on-demand chemical libraries can access far greater chemical space, provided that the predictive accuracy is sufficient to identify useful molecules. Through the largest and most diverse virtual HTS campaign reported to date, comprising 318 individual projects, we demonstrate that our AtomNet® convolutional neural network successfully finds novel hits across every major therapeutic area and protein class. We address historical limitations of computational screening by demonstrating success for target proteins without known binders, high-quality X-ray crystal structures, or manual cherry-picking of compounds. We show that the molecules selected by the AtomNet® model are novel drug-like scaffolds rather than minor modifications to known bioactive compounds. Our empirical results suggest that computational methods can substantially replace HTS as the first step of small-molecule drug discovery.
© 2024. The Author(s).
Conflict of interest statement
The authors affiliated with Atomwise declare the existence of a financial competing interest.
Figures


References
-
- Kuntz, I. D. Structure-based strategies for drug design and discovery. Science257, 1078–1082 (1992). - PubMed
-
- Bajorath, J. Integration of virtual and high-throughput screening. Nat. Rev. Drug Discov.1, 882–894 (2002). - PubMed
-
- Walters, W. P., Stahl, M. T. & Murcko, M. A. Virtual screening—an overview. Drug Discov. Today3, 160–178 (1998).
MeSH terms
Substances
Grants and funding
- R35 GM139817/GM/NIGMS NIH HHS/United States
- R01 AG076051/AG/NIA NIH HHS/United States
- R01 DK066485/DK/NIDDK NIH HHS/United States
- R01 HL165797/HL/NHLBI NIH HHS/United States
- R01 GM107129/GM/NIGMS NIH HHS/United States
- R01 AG065240/AG/NIA NIH HHS/United States
- R01 CA238229/CA/NCI NIH HHS/United States
- R01 DK083527/DK/NIDDK NIH HHS/United States
- R35 GM141849/GM/NIGMS NIH HHS/United States
- R01 AG064188/AG/NIA NIH HHS/United States
- R01 NS116055/NS/NINDS NIH HHS/United States
- R01 HL141432/HL/NHLBI NIH HHS/United States
- R01 CA264320/CA/NCI NIH HHS/United States
- R21 AI154088/AI/NIAID NIH HHS/United States
- UH2 CA223670/CA/NCI NIH HHS/United States
- R35 GM140852/GM/NIGMS NIH HHS/United States
- K08 DE028946/DE/NIDCR NIH HHS/United States
- R01 DK125263/DK/NIDDK NIH HHS/United States
- R01 CA260629/CA/NCI NIH HHS/United States
- R37 GM041376/GM/NIGMS NIH HHS/United States
- R01 AI136836/AI/NIAID NIH HHS/United States
- P50 CA150964/CA/NCI NIH HHS/United States
- R01 HL141353/HL/NHLBI NIH HHS/United States
- R01 AI157046/AI/NIAID NIH HHS/United States
- R35 GM140947/GM/NIGMS NIH HHS/United States
- R01 DK118940/DK/NIDDK NIH HHS/United States
- RF1 AG074346/AG/NIA NIH HHS/United States
- P51 OD011104/OD/NIH HHS/United States
- R01 AI147325/AI/NIAID NIH HHS/United States
- R01 GM099836/GM/NIGMS NIH HHS/United States
- R37 CA248565/CA/NCI NIH HHS/United States
- R01 AI161363/AI/NIAID NIH HHS/United States
- R01 EY025641/EY/NEI NIH HHS/United States
- R01 CA196643/CA/NCI NIH HHS/United States
- R35 GM133769/GM/NIGMS NIH HHS/United States
- R01 NS115903/NS/NINDS NIH HHS/United States
- R01 CA256791/CA/NCI NIH HHS/United States
- R01 DA048153/DA/NIDA NIH HHS/United States
- R01 AI179926/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources

