Characterization of a putative orexin receptor in Ciona intestinalis sheds light on the evolution of the orexin/hypocretin system in chordates
- PMID: 38565870
- PMCID: PMC10987541
- DOI: 10.1038/s41598-024-56508-1
Characterization of a putative orexin receptor in Ciona intestinalis sheds light on the evolution of the orexin/hypocretin system in chordates
Abstract
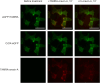
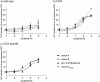
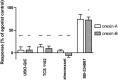
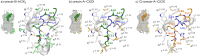
Tunicates are evolutionary model organisms bridging the gap between vertebrates and invertebrates. A genomic sequence in Ciona intestinalis (CiOX) shows high similarity to vertebrate orexin receptors and protostome allatotropin receptors (ATR). Here, molecular phylogeny suggested that CiOX is divergent from ATRs and human orexin receptors (hOX1/2). However, CiOX appears closer to hOX1/2 than to ATR both in terms of sequence percent identity and in its modelled binding cavity, as suggested by molecular modelling. CiOX was heterologously expressed in a recombinant HEK293 cell system. Human orexins weakly but concentration-dependently activated its Gq signalling (Ca2+ elevation), and the responses were inhibited by the non-selective orexin receptor antagonists TCS 1102 and almorexant, but only weakly by the OX1-selective antagonist SB-334867. Furthermore, the 5-/6-carboxytetramethylrhodamine (TAMRA)-labelled human orexin-A was able to bind to CiOX. Database mining was used to predict a potential endogenous C. intestinalis orexin peptide (Ci-orexin-A). Ci-orexin-A was able to displace TAMRA-orexin-A, but not to induce any calcium response at the CiOX. Consequently, we suggested that the orexin signalling system is conserved in Ciona intestinalis, although the relevant peptide-receptor interaction was not fully elucidated.
Keywords: Ciona intestinalis; Calcium; Hypocretin; Orexin; Receptor binding.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



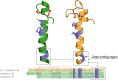
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

