Cellular metabolism regulates the differentiation and function of T-cell subsets
- PMID: 38565887
- PMCID: PMC11061161
- DOI: 10.1038/s41423-024-01148-8
Cellular metabolism regulates the differentiation and function of T-cell subsets
Abstract
T cells are an important component of adaptive immunity and protect the host from infectious diseases and cancers. However, uncontrolled T cell immunity may cause autoimmune disorders. In both situations, antigen-specific T cells undergo clonal expansion upon the engagement and activation of antigens. Cellular metabolism is reprogrammed to meet the increase in bioenergetic and biosynthetic demands associated with effector T cell expansion. Metabolites not only serve as building blocks or energy sources to fuel cell growth and expansion but also regulate a broad spectrum of cellular signals that instruct the differentiation of multiple T cell subsets. The realm of immunometabolism research is undergoing swift advancements. Encapsulating all the recent progress within this concise review in not possible. Instead, our objective is to provide a succinct introduction to this swiftly progressing research, concentrating on the metabolic intricacies of three pivotal nutrient classes-lipids, glucose, and amino acids-in T cells. We shed light on recent investigations elucidating the roles of these three groups of metabolites in mediating the metabolic and immune functions of T cells. Moreover, we delve into the prospect of "editing" metabolic pathways within T cells using pharmacological or genetic approaches, with the aim of synergizing this approach with existing immunotherapies and enhancing the efficacy of antitumor and antiinfection immune responses.
Keywords: CD4+ T cells; CD8+T cells; Immunometabolism; Metabolism; T cell differentiation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
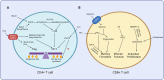
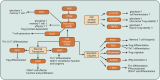
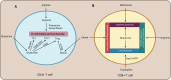
References
-
- O’Sullivan D, Pearce EL. Expanding the role of metabolism in T cells. Science. 2015;348:976–7. - PubMed
-
- Balmer ML, Ma EH, Bantug GR, Grahlert J, Pfister S, Glatter T, et al. Memory CD8(+) T cells require increased concentrations of acetate induced by stress for optimal function. Immunity. 2016;44:1312–24. - PubMed
-
- Berod L, Friedrich C, Nandan A, Freitag J, Hagemann S, Harmrolfs K, et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat Med. 2014;20:1327–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

