Direct and selective pharmacological disruption of the YAP-TEAD interface by IAG933 inhibits Hippo-dependent and RAS-MAPK-altered cancers
- PMID: 38565920
- PMCID: PMC11286534
- DOI: 10.1038/s43018-024-00754-9
Direct and selective pharmacological disruption of the YAP-TEAD interface by IAG933 inhibits Hippo-dependent and RAS-MAPK-altered cancers
Erratum in
-
Author Correction: Direct and selective pharmacological disruption of the YAP-TEAD interface by IAG933 inhibits Hippo-dependent and RAS-MAPK-altered cancers.Nat Cancer. 2024 Jul;5(7):1130. doi: 10.1038/s43018-024-00797-y. Nat Cancer. 2024. PMID: 38886525 Free PMC article. No abstract available.
Abstract
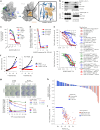
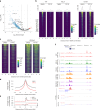
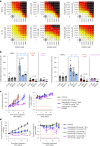
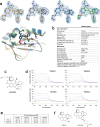
The YAP-TEAD protein-protein interaction mediates YAP oncogenic functions downstream of the Hippo pathway. To date, available YAP-TEAD pharmacologic agents bind into the lipid pocket of TEAD, targeting the interaction indirectly via allosteric changes. However, the consequences of a direct pharmacological disruption of the interface between YAP and TEADs remain largely unexplored. Here, we present IAG933 and its analogs as potent first-in-class and selective disruptors of the YAP-TEAD protein-protein interaction with suitable properties to enter clinical trials. Pharmacologic abrogation of the interaction with all four TEAD paralogs resulted in YAP eviction from chromatin and reduced Hippo-mediated transcription and induction of cell death. In vivo, deep tumor regression was observed in Hippo-driven mesothelioma xenografts at tolerated doses in animal models as well as in Hippo-altered cancer models outside mesothelioma. Importantly this also extended to larger tumor indications, such as lung, pancreatic and colorectal cancer, in combination with RTK, KRAS-mutant selective and MAPK inhibitors, leading to more efficacious and durable responses. Clinical evaluation of IAG933 is underway.
© 2024. The Author(s).
Conflict of interest statement
All authors are Novartis Pharma employees or were Novartis Pharma employees at the time of the studies.
Figures











References
-
- Currey, L., Thor, S. & Piper, M. TEAD family transcription factors in development and disease. Development148, dev196675 (2021). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

