Insights for disease modeling from single-cell transcriptomics of iPSC-derived Ngn2-induced neurons and astrocytes across differentiation time and co-culture
- PMID: 38566045
- PMCID: PMC10985965
- DOI: 10.1186/s12915-024-01867-4
Insights for disease modeling from single-cell transcriptomics of iPSC-derived Ngn2-induced neurons and astrocytes across differentiation time and co-culture
Abstract
Background: Trans-differentiation of human-induced pluripotent stem cells into neurons via Ngn2-induction (hiPSC-N) has become an efficient system to quickly generate neurons a likely significant advance for disease modeling and in vitro assay development. Recent single-cell interrogation of Ngn2-induced neurons, however, has revealed some similarities to unexpected neuronal lineages. Similarly, a straightforward method to generate hiPSC-derived astrocytes (hiPSC-A) for the study of neuropsychiatric disorders has also been described.
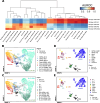
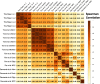
Results: Here, we examine the homogeneity and similarity of hiPSC-N and hiPSC-A to their in vivo counterparts, the impact of different lengths of time post Ngn2 induction on hiPSC-N (15 or 21 days), and the impact of hiPSC-N/hiPSC-A co-culture. Leveraging the wealth of existing public single-cell RNA-seq (scRNA-seq) data in Ngn2-induced neurons and in vivo data from the developing brain, we provide perspectives on the lineage origins and maturation of hiPSC-N and hiPSC-A. While induction protocols in different labs produce consistent cell type profiles, both hiPSC-N and hiPSC-A show significant heterogeneity and similarity to multiple in vivo cell fates, and both more precisely approximate their in vivo counterparts when co-cultured. Gene expression data from the hiPSC-N show enrichment of genes linked to schizophrenia (SZ) and autism spectrum disorders (ASD) as has been previously shown for neural stem cells and neurons. These overrepresentations of disease genes are strongest in our system at early times (day 15) in Ngn2-induction/maturation of neurons, when we also observe the greatest similarity to early in vivo excitatory neurons. We have assembled this new scRNA-seq data along with the public data explored here as an integrated biologist-friendly web-resource for researchers seeking to understand this system more deeply: https://nemoanalytics.org/p?l=DasEtAlNGN2&g=NES .
Conclusions: While overall we support the use of the investigated cellular models for the study of neuropsychiatric disease, we also identify important limitations. We hope that this work will contribute to understanding and optimizing cellular modeling for complex brain disorders.
Keywords: Alzheimer’s; Astrocytes; Autism; Induced pluripotent stem cells; Neurons; Ngn2; Schizophrenia; Single cell; Transcriptome.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Kerr CL, Letzen BS, Hill CM, Agrawal G, Thakor NV, Sterneckert JL, Gearhart JD, All AH. Efficient differentiation of human embryonic stem cells into oligodendrocyte progenitors for application in a rat contusion model of spinal cord injury. Int J Neurosci. 2010;120(4):305–313. doi: 10.3109/00207450903585290. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

