Single cell analysis reveals the roles and regulatory mechanisms of type-I interferons in Parkinson's disease
- PMID: 38566100
- PMCID: PMC10985960
- DOI: 10.1186/s12964-024-01590-1
Single cell analysis reveals the roles and regulatory mechanisms of type-I interferons in Parkinson's disease
Abstract
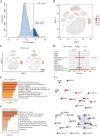
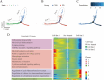
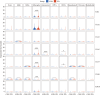
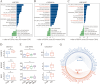
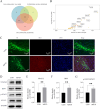
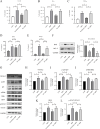
The pathogenesis of Parkinson's disease (PD) is strongly associated with neuroinflammation, and type I interferons (IFN-I) play a crucial role in regulating immune and inflammatory responses. However, the specific features of IFN in different cell types and the underlying mechanisms of PD have yet to be fully described. In this study, we analyzed the GSE157783 dataset, which includes 39,024 single-cell RNA sequencing results for five PD patients and six healthy controls from the Gene Expression Omnibus database. After cell type annotation, we intersected differentially expressed genes in each cell subcluster with genes collected in The Interferome database to generate an IFN-I-stimulated gene set (ISGs). Based on this gene set, we used the R package AUCell to score each cell, representing the IFN-I activity. Additionally, we performed monocle trajectory analysis, and single-cell regulatory network inference and clustering (SCENIC) to uncover the underlying mechanisms. In silico gene perturbation and subsequent experiments confirm NFATc2 regulation of type I interferon response and neuroinflammation. Our analysis revealed that microglia, endothelial cells, and pericytes exhibited the highest activity of IFN-I. Furthermore, single-cell trajectory detection demonstrated that microglia in the midbrain of PD patients were in a pro-inflammatory activation state, which was validated in the 1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced PD mouse model as well. We identified transcription factors NFATc2, which was significantly up-regulated and involved in the expression of ISGs and activation of microglia in PD. In the 1-Methyl-4-phenylpyridinium (MPP+)-induced BV2 cell model, the suppression of NFATc2 resulted in a reduction in IFN-β levels, impeding the phosphorylation of STAT1, and attenuating the activation of the NF-κB pathway. Furthermore, the downregulation of NFATc2 mitigated the detrimental effects on SH-SY5Y cells co-cultured in conditioned medium. Our study highlights the critical role of microglia in type I interferon responses in PD. Additionally, we identified transcription factors NFATc2 as key regulators of aberrant type I interferon responses and microglial pro-inflammatory activation in PD. These findings provide new insights into the pathogenesis of PD and may have implications for the development of novel therapeutic strategies.
Keywords: Immune; Parkinson’s disease; Single cell; Transcription factors; Type-I interferons.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

