ATF6 supports lysosomal function in tumor cells to enable ER stress-activated macroautophagy and CMA: impact on mutant TP53 expression
- PMID: 38566314
- PMCID: PMC11262222
- DOI: 10.1080/15548627.2024.2338577
ATF6 supports lysosomal function in tumor cells to enable ER stress-activated macroautophagy and CMA: impact on mutant TP53 expression
Abstract
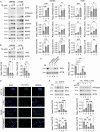
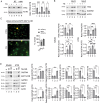
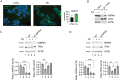
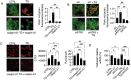
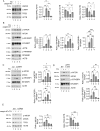
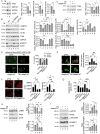
The inhibition of the unfolded protein response (UPR), which usually protects cancer cells from stress, may be exploited to potentiate the cytotoxic effect of drugs inducing ER stress. However, in this study, we found that ER stress and UPR activation by thapsigargin or tunicamycin promoted the lysosomal degradation of mutant (MUT) TP53 and that the inhibition of the UPR sensor ATF6, but not of ERN1/IRE1 or EIF2AK3/PERK, counteracted such an effect. ATF6 activation was indeed required to sustain the function of lysosomes, enabling the execution of chaperone-mediated autophagy (CMA) as well as of macroautophagy, processes involved in the degradation of MUT TP53 in stressed cancer cells. At the molecular level, by pharmacological and genetic approaches, we demonstrated that the inhibition of ATF6 correlated with the activation of MTOR and with TFEB and LAMP1 downregulation in thapsigargin-treated MUT TP53 carrying cells. We hypothesize that the rescue of MUT TP53 expression by ATF6 inhibition, could further activate MTOR and maintain lysosomal dysfunction, further inhibiting MUT TP53 degradation, in a vicious circle. The findings of this study suggest that the presence of MUT TP53, which often exerts oncogenic properties, should be considered before approaching treatments combining ER stressors with ATF6 inhibitors against cancer cells, while it could represent a promising strategy against cancer cells that harbor WT TP53.
Keywords: ATF6; CMA; UPR; cathepsins; mutant TP53; thapsigargin.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
Similar articles
-
Phosphorylation of EIF2S1 (eukaryotic translation initiation factor 2 subunit alpha) is indispensable for nuclear translocation of TFEB and TFE3 during ER stress.Autophagy. 2023 Jul;19(7):2111-2142. doi: 10.1080/15548627.2023.2173900. Epub 2023 Feb 9. Autophagy. 2023. PMID: 36719671 Free PMC article.
-
The unfolded protein response regulates hepatic autophagy by sXBP1-mediated activation of TFEB.Autophagy. 2021 Aug;17(8):1841-1855. doi: 10.1080/15548627.2020.1788889. Epub 2020 Jul 15. Autophagy. 2021. PMID: 32597296 Free PMC article.
-
RIPK1 regulates survival of human melanoma cells upon endoplasmic reticulum stress through autophagy.Autophagy. 2015;11(7):975-94. doi: 10.1080/15548627.2015.1049800. Autophagy. 2015. PMID: 26018731 Free PMC article.
-
Targeting UPR branches, a potential strategy for enhancing efficacy of cancer chemotherapy.Acta Biochim Biophys Sin (Shanghai). 2021 Nov 10;53(11):1417-1427. doi: 10.1093/abbs/gmab131. Acta Biochim Biophys Sin (Shanghai). 2021. PMID: 34664059 Review.
-
Unfolding anti-tumor immunity: ER stress responses sculpt tolerogenic myeloid cells in cancer.J Immunother Cancer. 2017 Jan 17;5:5. doi: 10.1186/s40425-016-0203-4. eCollection 2017. J Immunother Cancer. 2017. PMID: 28105371 Free PMC article. Review.
Cited by
-
Post-Translational Modifications (PTMs) of mutp53 and Epigenetic Changes Induced by mutp53.Biology (Basel). 2024 Jul 8;13(7):508. doi: 10.3390/biology13070508. Biology (Basel). 2024. PMID: 39056701 Free PMC article. Review.
-
The Role of ER Stress and the Unfolded Protein Response in Cancer.Cancer Genomics Proteomics. 2025 May-Jun;22(3):363-381. doi: 10.21873/cgp.20507. Cancer Genomics Proteomics. 2025. PMID: 40280715 Free PMC article. Review.
-
The role of autophagy in fibrosis: Mechanisms, progression and therapeutic potential (Review).Int J Mol Med. 2025 Apr;55(4):61. doi: 10.3892/ijmm.2025.5502. Epub 2025 Feb 14. Int J Mol Med. 2025. PMID: 39950330 Free PMC article. Review.
-
Mutant p53 upregulates HDAC6 to resist ER stress and facilitates Ku70 deacetylation, which prevents its degradation and mitigates DNA damage in colon cancer cells.Cell Death Discov. 2025 Apr 10;11(1):162. doi: 10.1038/s41420-025-02433-9. Cell Death Discov. 2025. PMID: 40210861 Free PMC article.
-
Recent Advances on Mutant p53: Unveiling Novel Oncogenic Roles, Degradation Pathways, and Therapeutic Interventions.Biomolecules. 2024 May 31;14(6):649. doi: 10.3390/biom14060649. Biomolecules. 2024. PMID: 38927053 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
