Single-cell analysis reveals a unique microenvironment in peri-implantitis
- PMID: 38566468
- PMCID: PMC11651718
- DOI: 10.1111/jcpe.13982
Single-cell analysis reveals a unique microenvironment in peri-implantitis
Abstract
Aim: This study aimed to reveal the unique microenvironment of peri-implantitis through single-cell analysis.
Materials and methods: Herein, we performed single-cell RNA sequencing (scRNA-seq) of biopsies from patients with peri-implantitis (PI) and compared the results with healthy individuals (H) and patients with periodontitis (PD).
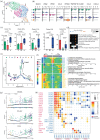
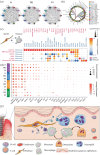
Results: Decreased numbers of stromal cells and increased immune cells were found in the PI group, which implies a severe inflammatory infiltration. The fibroblasts were found to be heterogeneous and the specific pro-inflammatory CXCL13+ sub-cluster was more represented in the PI group, in contrast to the PD and H groups. Furthermore, more neutrophil infiltration was detected in the PI group than in the PD group, and cell-cell communication and ligand-receptor pairs revealed most neutrophils were recruited by CXCL13+ fibroblasts through CXCL8/CXCL6-CXCR2/CXCR1. Notably, our study demonstrated that the unique microenvironment of the PI group promoted the differentiation of monocyte/macrophage lineage cells into osteoclasts, which might explain the faster and more severe bone resorption in the progression of PI than PD.
Conclusions: Collectively, this study suggests a unique immune microenvironment of PI, which may explain the differences between PI and PD in the clinic. These outcomes will aid in finding new specific and effective treatments for PI.
Keywords: immune microenvironment; periodontitis; peri‐implantitis; single‐cell RNA‐seq.
© 2024 The Authors. Journal of Clinical Periodontology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Berglundh, T. , Armitage, G. , Araujo, M. G. , Avila‐Ortiz, G. , Blanco, J. , Camargo, P. M. , Chen, S. , Cochran, D. , Derks, J. , Figuero, E. , & Hämmerle, C. H. (2018). Peri‐implant diseases and conditions: Consensus report of workgroup 4 of the 2017 World Workshop on the Classification of Periodontal and Peri‐Implant Diseases and Conditions. Journal of Periodontology, 89(Suppl 1), S313–s318. - PubMed
-
- Berglundh, T. , Zitzmann, N. U. , & Donati, M. (2011). Are peri‐implantitis lesions different from periodontitis lesions? Journal of Clinical Periodontology, 38(Suppl 11), 188–202. - PubMed
-
- Carcuac, O. , Abrahamsson, I. , Albouy, J. P. , Linder, E. , Larsson, L. , & Berglundh, T. (2013). Experimental periodontitis and peri‐implantitis in dogs. Clinical Oral Implants Research, 24, 363–371. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 824MS152/Hainan Provincial Natural Science Foundation of China
- YG2021QN72/Jiao Tong University Star" Program of Shanghai Jiao Tong University
- 2019-I2M-5-037/CAMS Innovation Fund for Medical Sciences (CIFMS)
- 82271004/National Natural Science Foundation of China
- KQYJXK2020/Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

