Unconventional modes of peptide-HLA-I presentation change the rules of TCR engagement
- PMID: 38566908
- PMCID: PMC10917088
- DOI: 10.1093/discim/kyac001
Unconventional modes of peptide-HLA-I presentation change the rules of TCR engagement
Abstract
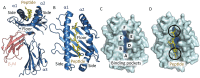
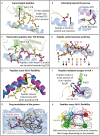
The intracellular proteome of virtually every nucleated cell in the body is continuously presented at the cell surface via the human leukocyte antigen class I (HLA-I) antigen processing pathway. This pathway classically involves proteasomal degradation of intracellular proteins into short peptides that can be presented by HLA-I molecules for interrogation by T-cell receptors (TCRs) expressed on the surface of CD8+ T cells. During the initiation of a T-cell immune response, the TCR acts as the T cell's primary sensor, using flexible loops to mould around the surface of the pHLA-I molecule to identify foreign or dysregulated antigens. Recent findings demonstrate that pHLA-I molecules can also be highly flexible and dynamic, altering their shape according to minor polymorphisms between different HLA-I alleles, or interactions with different peptides. These flexible presentation modes have important biological consequences that can, for example, explain why some HLA-I alleles offer greater protection against HIV, or why some cancer vaccine approaches have been ineffective. This review explores how these recent findings redefine the rules for peptide presentation by HLA-I molecules and extend our understanding of the molecular mechanisms that govern TCR-mediated antigen discrimination.
Keywords: T cells; antigen recognition; computational simulations; human leukocyte antigen (HLA); peptide presentation; protein flexibility.
© The Author(s) 2022. Published by Oxford University Press on behalf of the British Society for Immunology.
Figures
References
-
- Gorer PA. The detection of a hereditary antigenic difference in the blood of mice by means of human group a serum. J Genet 1936, 32, 17–31.
-
- Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, et al. Gene map of the extended human MHC. Nat Rev Genet 2004, 5, 889–99. - PubMed
-
- Rothbard JB, Gefter ML.. Interactions between immunogenic peptides and MHC proteins. Annu Rev Immunol 1991, 9, 527–65. - PubMed
-
- Guillet J-G, Lai M-Z, Briner TJ, Smith JA, Gefter ML.. Interaction of peptide antigens and class II major histocompatibility complex antigens. Nature 1986, 324, 260–2. - PubMed
-
- Davis MM, Bjorkman PJ.. The T cell receptor genes and T cell recognition. Nature 1988, 334, 395–402. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
