Combination of metagenomic next-generation sequencing and conventional tests unraveled pathogen profiles in infected patients undergoing allogeneic hematopoietic stem cell transplantation in Jilin Province of China
- PMID: 38567023
- PMCID: PMC10985322
- DOI: 10.3389/fcimb.2024.1378112
Combination of metagenomic next-generation sequencing and conventional tests unraveled pathogen profiles in infected patients undergoing allogeneic hematopoietic stem cell transplantation in Jilin Province of China
Abstract
Background: Infection is the main cause of death for patients after allogeneic hematopoietic stem cell transplantation (HSCT). However, pathogen profiles still have not been reported in detail due to their heterogeneity caused by geographic region.
Objective: To evaluate the performance of metagenomic next-generation sequencing (mNGS) and summarize regional pathogen profiles of infected patients after HSCT.
Methods: From February 2021 to August 2022, 64 patients, admitted to the Department of Hematology of The First Hospital of Jilin University for HSCT and diagnosed as suspected infections, were retrospectively enrolled.
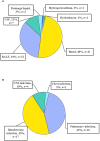
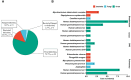
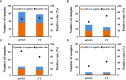
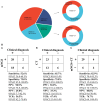
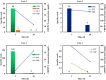
Results: A total of 38 patients were diagnosed as having infections, including bloodstream (n =17), pulmonary (n =16), central nervous system (CNS) (n =4), and chest (n =1) infections. Human betaherpesvirus 5 (CMV) was the most common pathogen in both bloodstream (n =10) and pulmonary (n =8) infections, while CNS (n =2) and chest (n =1) infections were mainly caused by Human gammaherpesvirus 4 (EBV). For bloodstream infection, Mycobacterium tuberculosis complex (n =3), Staphylococcus epidermidis (n =1), and Candida tropicalis (n =1) were also diagnosed as causative pathogens. Furthermore, mNGS combined with conventional tests can identify more causative pathogens with high sensitivity of 82.9% (95% CI 70.4-95.3%), and the total coincidence rate can reach up to 76.7% (95% CI 64.1-89.4%).
Conclusions: Our findings emphasized the importance of mNGS in diagnosing, managing, and ruling out infections, and an era of more rapid, independent, and impartial diagnosis of infections after HSCT can be expected.
Keywords: Jilin Province; MNGs; allogeneic hematopoietic stem cell transplantation; pathogen profiles; total coincidence rate.
Copyright © 2024 Zou, Gao, Liu, Liu, Xiao, Li and Jiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Metagenomic Next-Generation Sequencing in the Diagnosis of Pulmonary Infections after Allogeneic Hematopoietic Stem Cell Transplantation.Transplant Cell Ther. 2024 Nov;30(11):1110.e1-1110.e10. doi: 10.1016/j.jtct.2024.08.014. Epub 2024 Aug 27. Transplant Cell Ther. 2024. PMID: 39197493
-
Metagenomic Next-Generation Sequencing vs. Traditional Pathogen Detection in the Diagnosis of Infection After Allogeneic Hematopoietic Stem Cell Transplantation in Children.Front Microbiol. 2022 Apr 18;13:868160. doi: 10.3389/fmicb.2022.868160. eCollection 2022. Front Microbiol. 2022. PMID: 35509305 Free PMC article.
-
[Metagenomic next-generation sequencing for the diagnosis of Pneumocystis jirovecii pneumonia after allogeneic hematopoietic stem cell transplantation].Zhonghua Xue Ye Xue Za Zhi. 2024 Jan 14;45(1):62-67. doi: 10.3760/cma.j.cn121090-20230928-00147. Zhonghua Xue Ye Xue Za Zhi. 2024. PMID: 38527840 Free PMC article. Chinese.
-
Metagenomic Next-Generation Sequencing Contributes to the Early Diagnosis of Mixed Infections in Central Nervous System.Mycopathologia. 2024 Apr 18;189(3):34. doi: 10.1007/s11046-024-00837-2. Mycopathologia. 2024. PMID: 38637353 Review.
-
Rapid diagnosis of Talaromyces marneffei infection by metagenomic next-generation sequencing technology in a Chinese cohort of inborn errors of immunity.Front Cell Infect Microbiol. 2022 Sep 8;12:987692. doi: 10.3389/fcimb.2022.987692. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36159645 Free PMC article.
Cited by
-
Application of targeted metagenomic next-generation sequencing in pneumonia patients.Microbiol Spectr. 2025 Aug 5;13(8):e0171324. doi: 10.1128/spectrum.01713-24. Epub 2025 Jun 23. Microbiol Spectr. 2025. PMID: 40548737 Free PMC article.
References
-
- Ai J. W., Zhang H. C., Cui P., Xu B., Gao Y., Cheng Q., et al. . (2018). Dynamic and direct pathogen load surveillance to monitor disease progression and therapeutic efficacy in central nervous system infection using a novel semi-quantitive sequencing platform. J. Infect. 76, 307–310. doi: 10.1016/j.jinf.2017.11.002 - DOI - PubMed
-
- Berhane M., Gidi N. W., Eshetu B., Gashaw M., Tesfaw G., Wieser A., et al. . (2021). Clinical profile of neonates admitted with sepsis to neonatal intensive care unit of Jimma Medical Center, a tertiary hospital in Ethiopia. Ethiopian J. Health Sci. 31, 485– 494. doi: 10.4314/ejhs.v31i3.5 - DOI - PMC - PubMed
-
- Chen Y., Feng W., Ye K., Guo L., Xia H., Guan Y., et al. . (2021). Application of metagenomic next-generation sequencing in the diagnosis of pulmonary infectious pathogens from bronchoalveolar lavage samples. Front. Cell Infect. Microbiol. 11, 168. doi: 10.3389/fcimb.2021.541092 - DOI - PMC - PubMed
-
- Chen H., Yin Y., Gao H., Guo Y., Dong Z., Wang X., et al. . (2020). Clinical utility of in-house metagenomic next-generation sequencing for the diagnosis of lower respiratory tract infections and analysis of the host immune response. Clin. Infect. Dis. 71, S416–S426. doi: 10.1093/cid/ciaa1516 - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

