Revisiting Exome Data Identified Missed Splice Site Variant of the Asparagine Synthetase ( ASNS ) Gene
- PMID: 38567172
- PMCID: PMC10984708
- DOI: 10.1055/s-0042-1757193
Revisiting Exome Data Identified Missed Splice Site Variant of the Asparagine Synthetase ( ASNS ) Gene
Abstract
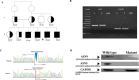
Next-generation sequencing, such as whole-exome sequencing (WES), is increasingly used in the study of Mendelian disorders, yet many are reported as "negative." Inappropriate variant annotation and filtering steps are reasons for missing the molecular diagnosis. Noncoding variants, including splicing mutations, are examples of variants that can be overlooked. Herein, we report a family of four affected newborns, and all presented with severe congenital microcephaly. Initial research WES analysis identified a damaging homozygous variant in NME1 gene as a possible cause of primary microcephaly phenotype in these patients. However, reanalysis of the exome data uncovered a biallelic splice site variant in asparagine synthetase gene which seems to be the possible cause of the phenotype in these patients. This study highlights the importance of revisiting the exome data and the issue of "negative" exome and the afterward approaches to identify and prove new candidate genes.
Keywords: ASNS; NME1; candidate genes; microcephaly; whole-exome sequencing.
Thieme. All rights reserved.
Conflict of interest statement
Conflict of Interest None declared.
Figures

Similar articles
-
Asparagine synthetase deficiency detected by whole exome sequencing causes congenital microcephaly, epileptic encephalopathy and psychomotor delay.Metab Brain Dis. 2015 Jun;30(3):687-94. doi: 10.1007/s11011-014-9618-0. Epub 2014 Sep 17. Metab Brain Dis. 2015. PMID: 25227173 Free PMC article.
-
Case Report: a novel homozygous ASNS variant in a Chinese female with severe microcephaly, encephalopathy and epilepsy.Front Neurosci. 2025 May 12;19:1570160. doi: 10.3389/fnins.2025.1570160. eCollection 2025. Front Neurosci. 2025. PMID: 40421135 Free PMC article.
-
Case report: A compound heterozygous mutations in ASNS broadens the spectrum of asparagine synthetase deficiency in the prenatal diagnosis.Front Pediatr. 2023 Oct 13;11:1273789. doi: 10.3389/fped.2023.1273789. eCollection 2023. Front Pediatr. 2023. PMID: 37900678 Free PMC article.
-
A novel variant in ASNS gene responsible for syndromic intellectual disability and microcephaly: Case report and literature review.Mol Genet Genomic Med. 2024 Apr;12(4):e2424. doi: 10.1002/mgg3.2424. Mol Genet Genomic Med. 2024. PMID: 38546112 Free PMC article. Review.
-
Added Value of Reanalysis of Whole Exome- and Whole Genome Sequencing Data From Patients Suspected of Primary Immune Deficiency Using an Extended Gene Panel and Structural Variation Calling.Front Immunol. 2022 Jun 30;13:906328. doi: 10.3389/fimmu.2022.906328. eCollection 2022. Front Immunol. 2022. PMID: 35874679 Free PMC article. Review.
References
-
- Bruel A. L. et al.Next-generation sequencing approaches and challenges in the diagnosis of developmental anomalies and intellectual disability. Clinical Genetics. 2020;98:433–444. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

