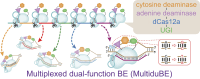
Multiplexed in-situ mutagenesis driven by a dCas12a-based dual-function base editor
- PMID: 38567723
- PMCID: PMC11077070
- DOI: 10.1093/nar/gkae228
Multiplexed in-situ mutagenesis driven by a dCas12a-based dual-function base editor
Abstract
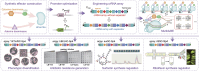
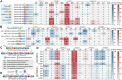
Mutagenesis driving genetic diversity is vital for understanding and engineering biological systems. However, the lack of effective methods to generate in-situ mutagenesis in multiple genomic loci combinatorially limits the study of complex biological functions. Here, we design and construct MultiduBE, a dCas12a-based multiplexed dual-function base editor, in an all-in-one plasmid for performing combinatorial in-situ mutagenesis. Two synthetic effectors, duBE-1a and duBE-2b, are created by amalgamating the functionalities of cytosine deaminase (from hAPOBEC3A or hAID*Δ ), adenine deaminase (from TadA9), and crRNA array processing (from dCas12a). Furthermore, introducing the synthetic separator Sp4 minimizes interference in the crRNA array, thereby facilitating multiplexed in-situ mutagenesis in both Escherichia coli and Bacillus subtilis. Guided by the corresponding crRNA arrays, MultiduBE is successfully employed for cell physiology reprogramming and metabolic regulation. A novel mutation conferring streptomycin resistance has been identified in B. subtilis and incorporated into the mutant strains with multiple antibiotic resistance. Moreover, surfactin and riboflavin titers of the combinatorially mutant strains improved by 42% and 15-fold, respectively, compared with the control strains with single gene mutation. Overall, MultiduBE provides a convenient and efficient way to perform multiplexed in-situ mutagenesis.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
CRISPR-Directed Gene Editing Catalyzes Precise Gene Segment Replacement In Vitro Enabling a Novel Method for Multiplex Site-Directed Mutagenesis.CRISPR J. 2019 Apr;2:121-132. doi: 10.1089/crispr.2018.0054. CRISPR J. 2019. PMID: 30998096
-
Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis.Nature. 2019 Jul;571(7764):275-278. doi: 10.1038/s41586-019-1314-0. Epub 2019 Jun 10. Nature. 2019. PMID: 31181567
-
CRISPR-Cas-Guided Mutagenesis of Chromosome and Virulence Plasmid in Shigella flexneri by Cytosine Base Editing.mSystems. 2023 Feb 23;8(1):e0104522. doi: 10.1128/msystems.01045-22. Epub 2022 Dec 21. mSystems. 2023. PMID: 36541764 Free PMC article.
-
Single-nucleotide editing: From principle, optimization to application.Hum Mutat. 2019 Dec;40(12):2171-2183. doi: 10.1002/humu.23819. Epub 2019 Sep 15. Hum Mutat. 2019. PMID: 31131955 Free PMC article. Review.
-
The CRISPR toolbox for the gram-positive model bacterium Bacillus subtilis.Crit Rev Biotechnol. 2022 Sep;42(6):813-826. doi: 10.1080/07388551.2021.1983516. Epub 2021 Oct 31. Crit Rev Biotechnol. 2022. PMID: 34719304 Review.
Cited by
-
Precision multiplexed base editing in human cells using Cas12a-derived base editors.Nat Commun. 2025 May 31;16(1):5061. doi: 10.1038/s41467-025-59653-x. Nat Commun. 2025. PMID: 40449999 Free PMC article.
-
T7 RNA polymerase-guided base editor for accelerated continuous evolution in Bacillus subtilis.Synth Syst Biotechnol. 2025 Apr 21;10(3):876-886. doi: 10.1016/j.synbio.2025.04.010. eCollection 2025 Sep. Synth Syst Biotechnol. 2025. PMID: 40386441 Free PMC article.
-
CRISPR/Cas13X-assisted programmable and multiplexed translation regulation for controlled biosynthesis.Nucleic Acids Res. 2025 Jan 7;53(1):gkae1293. doi: 10.1093/nar/gkae1293. Nucleic Acids Res. 2025. PMID: 39777467 Free PMC article.
-
A cross-species inducible system for enhanced protein expression and multiplexed metabolic pathway fine-tuning in bacteria.Nucleic Acids Res. 2025 Jan 11;53(2):gkae1315. doi: 10.1093/nar/gkae1315. Nucleic Acids Res. 2025. PMID: 39797735 Free PMC article.
References
-
- Korry B.J., Lee S.Y.E., Chakrabarti A.K., Choi A.H., Ganser C., Machan J.T., Belenky P.. Genotoxic agents produce stressor-specific spectra of spectinomycin resistance mutations based on mechanism of action and selection in Bacillus subtilis. Antimicrob. Agents Chemother. 2021; 65:e00891-21. - PMC - PubMed
-
- Halperin S.O., Tou C.J., Wong E.B., Modavi C., Schaffer D.V., Dueber J.E.. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature. 2018; 560:248–252. - PubMed
-
- Tian R., Rehm F.B.H., Czernecki D., Gu Y., Zürcher J.F., Liu K.C., Chin J.W.. Establishing a synthetic orthogonal replication system enables accelerated evolution in E. coli. Science. 2024; 383:421–426. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

