Vitamin D constrains inflammation by modulating the expression of key genes on Chr17q12-21.1
- PMID: 38567749
- PMCID: PMC10990493
- DOI: 10.7554/eLife.89270
Vitamin D constrains inflammation by modulating the expression of key genes on Chr17q12-21.1
Abstract
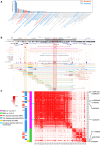
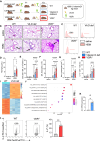
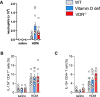
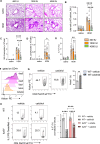
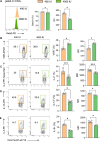
Vitamin D possesses immunomodulatory functions and vitamin D deficiency has been associated with the rise in chronic inflammatory diseases, including asthma (Litonjua and Weiss, 2007). Vitamin D supplementation studies do not provide insight into the molecular genetic mechanisms of vitamin D-mediated immunoregulation. Here, we provide evidence for vitamin D regulation of two human chromosomal loci, Chr17q12-21.1 and Chr17q21.2, reliably associated with autoimmune and chronic inflammatory diseases. We demonstrate increased vitamin D receptor (Vdr) expression in mouse lung CD4+ Th2 cells, differential expression of Chr17q12-21.1 and Chr17q21.2 genes in Th2 cells based on vitamin D status and identify the IL-2/Stat5 pathway as a target of vitamin D signaling. Vitamin D deficiency caused severe lung inflammation after allergen challenge in mice that was prevented by long-term prenatal vitamin D supplementation. Mechanistically, vitamin D induced the expression of the Ikzf3-encoded protein Aiolos to suppress IL-2 signaling and ameliorate cytokine production in Th2 cells. These translational findings demonstrate mechanisms for the immune protective effect of vitamin D in allergic lung inflammation with a strong molecular genetic link to the regulation of both Chr17q12-21.1 and Chr17q21.2 genes and suggest further functional studies and interventional strategies for long-term prevention of asthma and other autoimmune disorders.
Keywords: asthma; genetics; genomics; human; immunology; inflammation; mouse; vitamin D receptor.
© 2023, Kilic, Halu et al.
Conflict of interest statement
AK, AH, MD, EM, MD, TB, JR, RC, HM, AS, JK, TN, HP, NK, RA, KG, AL, MD, HR, BL No competing interests declared, SW receives royalties from UpToDate and is an investor in Histolix
Figures









Update of
- doi: 10.1101/2022.05.22.491886
- doi: 10.7554/eLife.89270.1
- doi: 10.7554/eLife.89270.2
- doi: 10.7554/eLife.89270.3
Comment in
- doi: 10.7554/eLife.97031
References
-
- Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, Chateigner N, Gormand F, Just J, Le Moual N, Scheinmann P, Siroux V, Vervloet D, Zelenika D, Pin I, Kauffmann F, Lathrop M, Demenais F. Effect of 17q21 variants and smoking exposure in early-onset asthma. The New England Journal of Medicine. 2008;359:1985–1994. doi: 10.1056/NEJMoa0806604. - DOI - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- K01 HL166705/HL/NHLBI NIH HHS/United States
- 5K25HL150336/HL/NHLBI NIH HHS/United States
- R01 HL091528/HL/NHLBI NIH HHS/United States
- R01 HL171141/HL/NHLBI NIH HHS/United States
- P30 ES001247/ES/NIEHS NIH HHS/United States
- K25 HL168157/HL/NHLBI NIH HHS/United States
- 1K01HL166705/HL/NHLBI NIH HHS/United States
- R01 HL122531/HL/NHLBI NIH HHS/United States
- 1R01HL171141/HL/NHLBI NIH HHS/United States
- K01 HL146977/HL/NHLBI NIH HHS/United States
- 1K25HL168157/HL/NHLBI NIH HHS/United States
- K25 HL150336/HL/NHLBI NIH HHS/United States
- 5P01HL132825/HL/NHLBI NIH HHS/United States
- 5UH3OD023268/HL/NHLBI NIH HHS/United States
- P01 HL132825/HL/NHLBI NIH HHS/United States
- UH3 OD023268/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

