Adaptive response of Pseudomonas aeruginosa under serial ciprofloxacin exposure
- PMID: 38568202
- PMCID: PMC11084610
- DOI: 10.1099/mic.0.001443
Adaptive response of Pseudomonas aeruginosa under serial ciprofloxacin exposure
Abstract
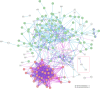
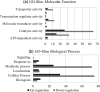
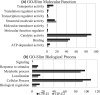
Understanding the evolution of antibiotic resistance is important for combating drug-resistant bacteria. In this work, we investigated the adaptive response of Pseudomonas aeruginosa to ciprofloxacin. Ciprofloxacin-susceptible P. aeruginosa ATCC 9027, CIP-E1 (P. aeruginosa ATCC 9027 exposed to ciprofloxacin for 14 days) and CIP-E2 (CIP-E1 cultured in antibiotic-free broth for 10 days) were compared. Phenotypic responses including cell morphology, antibiotic susceptibility, and production of pyoverdine, pyocyanin and rhamnolipid were assessed. Proteomic responses were evaluated using comparative iTRAQ labelling LC-MS/MS to identify differentially expressed proteins (DEPs). Expression of associated genes coding for notable DEPs and their related regulatory genes were checked using quantitative reverse transcriptase PCR. CIP-E1 displayed a heterogeneous morphology, featuring both filamentous cells and cells with reduced length and width. By contrast, although filaments were not present, CIP-E2 still exhibited size reduction. Considering the MIC values, ciprofloxacin-exposed strains developed resistance to fluoroquinolone antibiotics but maintained susceptibility to other antibiotic classes, except for carbapenems. Pyoverdine and pyocyanin production showed insignificant decreases, whereas there was a significant decrease in rhamnolipid production. A total of 1039 proteins were identified, of which approximately 25 % were DEPs. In general, there were more downregulated proteins than upregulated proteins. Noted changes included decreased OprD and PilP, and increased MexEF-OprN, MvaT and Vfr, as well as proteins of ribosome machinery and metabolism clusters. Gene expression analysis confirmed the proteomic data and indicated the downregulation of rpoB and rpoS. In summary, the response to CIP involved approximately a quarter of the proteome, primarily associated with ribosome machinery and metabolic processes. Potential targets for bacterial interference encompassed outer membrane proteins and global regulators, such as MvaT.
Keywords: Pseudomonas aeruginosa; antibiotic resistance; ciprofloxacin; iTraq labelling; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

