SF3A2 promotes progression and cisplatin resistance in triple-negative breast cancer via alternative splicing of MKRN1
- PMID: 38569025
- PMCID: PMC10990288
- DOI: 10.1126/sciadv.adj4009
SF3A2 promotes progression and cisplatin resistance in triple-negative breast cancer via alternative splicing of MKRN1
Abstract
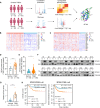
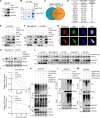
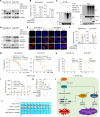
Triple-negative breast cancer (TNBC) is the deadliest subtype of breast cancer owing to the lack of effective therapeutic targets. Splicing factor 3a subunit 2 (SF3A2), a poorly defined splicing factor, was notably elevated in TNBC tissues and promoted TNBC progression, as confirmed by cell proliferation, colony formation, transwell migration, and invasion assays. Mechanistic investigations revealed that E3 ubiquitin-protein ligase UBR5 promoted the ubiquitination-dependent degradation of SF3A2, which in turn regulated UBR5, thus forming a feedback loop to balance these two oncoproteins. Moreover, SF3A2 accelerated TNBC progression by, at least in part, specifically regulating the alternative splicing of makorin ring finger protein 1 (MKRN1) and promoting the expression of the dominant and oncogenic isoform, MKRN1-T1. Furthermore, SF3A2 participated in the regulation of both extrinsic and intrinsic apoptosis, leading to cisplatin resistance in TNBC cells. Collectively, these findings reveal a previously unknown role of SF3A2 in TNBC progression and cisplatin resistance, highlighting SF3A2 as a potential therapeutic target for patients with TNBC.
Figures




References
-
- Gluz O., Liedtke C., Gottschalk N., Pusztai L., Nitz U., Harbeck N., Triple-negative breast cancer—Current status and future directions. Ann. Oncol. 20, 1913–1927 (2009). - PubMed
-
- Billar J. A. Y., Dueck A. C., Stucky C.-C. H., Gray R. J., Wasif N., Northfelt D. W., McCullough A. E., Pockaj B. A., Triple-negative breast cancers: Unique clinical presentations and outcomes. Ann. Surg. Oncol. 17, 384–390 (2010). - PubMed
-
- Malorni L., Shetty P. B., De Angelis C., Hilsenbeck S., Rimawi M. F., Elledge R., Osborne C. K., De Placido S., Arpino G., Clinical and biologic features of triple-negative breast cancers in a large cohort of patients with long-term follow-up. Breast Cancer Res. Treat. 136, 795–804 (2012). - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

