The Notch-PDGFRβ axis suppresses brown adipocyte progenitor differentiation in early post-natal mice
- PMID: 38569546
- PMCID: PMC11874136
- DOI: 10.1016/j.devcel.2024.03.012
The Notch-PDGFRβ axis suppresses brown adipocyte progenitor differentiation in early post-natal mice
Abstract
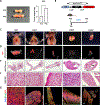
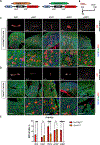
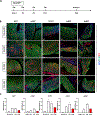
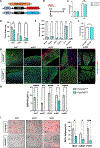
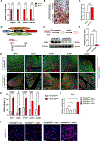
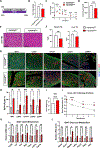
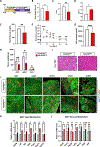
De novo brown adipogenesis holds potential in combating the epidemics of obesity and diabetes. However, the identity of brown adipocyte progenitor cells (APCs) and their regulation have not been extensively explored. Here, through in vivo lineage tracing and mouse modeling, we observed that platelet-derived growth factor receptor beta (PDGFRβ)+ pericytes give rise to developmental brown adipocytes but not to those in adult homeostasis. By contrast, T-box 18 (TBX18)+ pericytes contribute to brown adipogenesis throughout both developmental and adult stages, though in a depot-specific manner. Mechanistically, Notch inhibition in PDGFRβ+ pericytes promotes brown adipogenesis by downregulating PDGFRβ. Furthermore, inhibition of Notch signaling in PDGFRβ+ pericytes mitigates high-fat, high-sucrose (HFHS)-induced glucose and metabolic impairment in mice during their development and juvenile phases. Collectively, these findings show that the Notch/PDGFRβ axis negatively regulates developmental brown adipogenesis, and its repression promotes brown adipose tissue expansion and improves metabolic health.
Keywords: PDGFRβ; Tbx18; brown adipocyte progenitor cells; development; obesity; pericytes.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
Update of
-
The Notch-Pdgfrβ axis suppresses brown adipocyte progenitor differentiation in early postnatal mice.bioRxiv [Preprint]. 2023 May 24:2023.05.24.541839. doi: 10.1101/2023.05.24.541839. bioRxiv. 2023. Update in: Dev Cell. 2024 May 20;59(10):1233-1251.e5. doi: 10.1016/j.devcel.2024.03.012. PMID: 37293108 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

