Immune microniches shape intestinal Treg function
- PMID: 38570678
- PMCID: PMC11041794
- DOI: 10.1038/s41586-024-07251-0
Immune microniches shape intestinal Treg function
Abstract
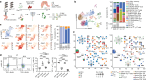
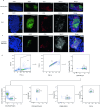
The intestinal immune system is highly adapted to maintaining tolerance to the commensal microbiota and self-antigens while defending against invading pathogens1,2. Recognizing how the diverse network of local cells establish homeostasis and maintains it in the complex immune environment of the gut is critical to understanding how tolerance can be re-established following dysfunction, such as in inflammatory disorders. Although cell and molecular interactions that control T regulatory (Treg) cell development and function have been identified3,4, less is known about the cellular neighbourhoods and spatial compartmentalization that shapes microorganism-reactive Treg cell function. Here we used in vivo live imaging, photo-activation-guided single-cell RNA sequencing5-7 and spatial transcriptomics to follow the natural history of T cells that are reactive towards Helicobacter hepaticus through space and time in the settings of tolerance and inflammation. Although antigen stimulation can occur anywhere in the tissue, the lamina propria-but not embedded lymphoid aggregates-is the key microniche that supports effector Treg (eTreg) cell function. eTreg cells are stable once their niche is established; however, unleashing inflammation breaks down compartmentalization, leading to dominance of CD103+SIRPα+ dendritic cells in the lamina propria. We identify and validate the putative tolerogenic interaction between CD206+ macrophages and eTreg cells in the lamina propria and identify receptor-ligand pairs that are likely to govern the interaction. Our results reveal a spatial mechanism of tolerance in the lamina propria and demonstrate how knowledge of local interactions may contribute to the next generation of tolerance-inducing therapies.
© 2024. The Author(s).
Conflict of interest statement
C.H. is a current employee of F. Hoffmann-La Roche. S.A.T. is a remunerated member of the scientific advisory board of Element Biosciences, Foresite Labs and Qiagen. S.A.T. is co-founder and equity holder of Transition Bio and EnsoCell, as well as a part-time employee of GlaxoSmithKline. F.P. receives consultancy fees or research support from Janssen, GSK, Genentech, and T-Cypher. The other authors declare no competing interests.
Figures












References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

