Deciphering the prognostic features of bladder cancer through gemcitabine resistance and immune-related gene analysis and identifying potential small molecular drug PIK-75
- PMID: 38570787
- PMCID: PMC10993528
- DOI: 10.1186/s12935-024-03258-9
Deciphering the prognostic features of bladder cancer through gemcitabine resistance and immune-related gene analysis and identifying potential small molecular drug PIK-75
Abstract
Background: Bladder cancer (BCa) stands out as a prevalent and highly lethal malignancy worldwide. Chemoresistance significantly contributes to cancer recurrence and progression. Traditional Tumor Node Metastasis (TNM) stage and molecular subtypes often fail to promptly identify treatment preferences based on sensitivity.
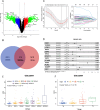
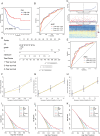
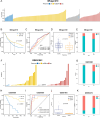
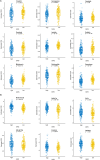
Methods: In this study, we developed a prognostic signature for BCa with uni-Cox + LASSO + multi-Cox survival analysis in multiple independent cohorts. Six machine learning algorithms were adopted to screen out the hub gene, RAC3. IHC staining was used to validate the expression of RAC3 in BCa tumor tissue. RT-qPCR and Western blot were performed to detect and quantify the mRNA and protein levels of RAC3. CCK8, colony formation, wound healing, and flow cytometry analysis of apoptosis were employed to determine cell proliferation, migration, and apoptosis. Molecular docking was used to find small target drugs, PIK-75. 3D cell viability assay was applied to evaluate the ATP viability of bladder cancer organoids before and after PIK-75 treated.
Results: The established clinical prognostic model, GIRS, comprises 13 genes associated with gemcitabine resistance and immunology. This model has demonstrated robust predictive capabilities for survival outcomes across various independent public cohorts. Additionally, the GIRS signature shows significant correlations with responses to both immunotherapy and chemotherapy. Leveraging machine learning algorithms, the hub gene, RAC3, was identified, and potential upstream transcription factors were screened through database analysis. IHC results showed that RAC3 was higher expressed in GEM-resistant BCa patients. Employing molecular docking, the small molecule drug PIK-75, as binding to RAC3, was identified. Experiments on cell lines, organoids and animals validated the biological effects of PIK-75 in bladder cancer.
Conclusions: The GIRS signature offers a valuable complement to the conventional anatomic TNM staging system and molecular subtype stratification in bladder cancer. The hub gene, RAC3, plays a crucial role in BCa and is significantly associated with resistance to gemcitabine. The small molecular drug, PIK-75 having the potential as a therapeutic agent in the context of gemcitabine-resistant and immune-related pathways.
Keywords: Bladder cancer; Machine learning; PIK-75; Prognostic signature; RAC3.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Babjuk M, Burger M, Capoun O, Cohen D, Compérat EM, Dominguez Escrig JL, Gontero P, Liedberg F, Masson-Lecomte A, Mostafid AH, et al. European association of urology guidelines on non-muscle-invasive bladder cancer (Ta, T1, and carcinoma in situ) Eur Urol. 2022;81:75–94. doi: 10.1016/j.eururo.2021.08.010. - DOI - PubMed
-
- Zou Y, Xie J, Zheng S, Liu W, Tang Y, Tian W, Deng X, Wu L, Zhang Y, Wong C-W, et al. Leveraging diverse cell-death patterns to predict the prognosis and drug sensitivity of triple-negative breast cancer patients after surgery. Int J Surg. 2022;107:106936. doi: 10.1016/j.ijsu.2022.106936. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

