Neuroligin-2 shapes individual slow waves during slow-wave sleep and the response to sleep deprivation in mice
- PMID: 38570872
- PMCID: PMC10993465
- DOI: 10.1186/s13229-024-00594-5
Neuroligin-2 shapes individual slow waves during slow-wave sleep and the response to sleep deprivation in mice
Abstract
Background: Sleep disturbances are a common comorbidity to most neurodevelopmental disorders and tend to worsen disease symptomatology. It is thus crucial to understand mechanisms underlying sleep disturbances to improve patients' quality of life. Neuroligin-2 (NLGN2) is a synaptic adhesion protein regulating GABAergic transmission. It has been linked to autism spectrum disorders and schizophrenia in humans, and deregulations of its expression were shown to cause epileptic-like hypersynchronized cerebral activity in rodents. Importantly, the absence of Nlgn2 (knockout: KO) was previously shown to alter sleep-wake duration and quality in mice, notably increasing slow-wave sleep (SWS) delta activity (1-4 Hz) and altering its 24-h dynamics. This type of brain oscillation is involved in memory consolidation, and is also a marker of homeostatic sleep pressure. Sleep deprivation (SD) is notably known to impair cognition and the physiological response to sleep loss involves GABAergic transmission.
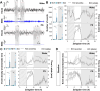
Methods: Using electrocorticographic (ECoG) recordings, we here first aimed to verify how individual slow wave (SW; 0.5-4 Hz) density and properties (e.g., amplitude, slope, frequency) contribute to the higher SWS delta activity and altered 24-h dynamics observed in Nlgn2 KO mice. We further investigated the response of these animals to SD. Finally, we tested whether sleep loss affects the gene expression of Nlgn2 and related GABAergic transcripts in the cerebral cortex of wild-type mice using RNA sequencing.
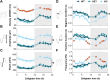
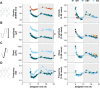
Results: Our results show that Nlgn2 KO mice have both greater SW amplitude and density, and that SW density is the main property contributing to the altered 24-h dynamics. We also found the absence of Nlgn2 to accelerate paradoxical sleep recovery following SD, together with profound alterations in ECoG activity across vigilance states. Sleep loss, however, did not modify the 24-h distribution of the hypersynchronized ECoG events observed in these mice. Finally, RNA sequencing confirmed an overall decrease in cortical expression of Nlgn2 and related GABAergic transcripts following SD in wild-type mice.
Conclusions: This work brings further insight into potential mechanisms of sleep duration and quality deregulation in neurodevelopmental disorders, notably involving NLGN2 and GABAergic neurotransmission.
Keywords: Cerebral cortex; GABAergic neurotransmission; Gene expression; Mice; Sleep deprivation; Sleep-wake regulation; Slow waves; Synaptic adhesion molecules.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

