Screening for immune-related biomarkers associated with myasthenia gravis and dilated cardiomyopathy based on bioinformatics analysis and machine learning
- PMID: 38571624
- PMCID: PMC10988011
- DOI: 10.1016/j.heliyon.2024.e28446
Screening for immune-related biomarkers associated with myasthenia gravis and dilated cardiomyopathy based on bioinformatics analysis and machine learning
Abstract
Background: We aim to investigate genes associated with myasthenia gravis (MG), specifically those potentially implicated in the pathogenesis of dilated cardiomyopathy (DCM). Additionally, we seek to identify potential biomarkers for diagnosing myasthenia gravis co-occurring with DCM.
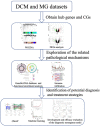
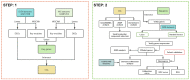
Methods: We obtained two expression profiling datasets related to DCM and MG from the Gene Expression Omnibus (GEO). Subsequently, we conducted differential gene expression analysis and weighted gene co-expression network analysis (WGCNA) on these datasets. The genes exhibiting differential expression common to both DCM and MG were employed for protein-protein interaction (PPI), Gene Ontology (GO) enrichment analysis, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Additionally, machine learning techniques were employed to identify potential biomarkers and develop a diagnostic nomogram for predicting MG-associated DCM. Subsequently, the machine learning results underwent validation using an external dataset. Finally, gene set enrichment analysis (GSEA) and machine algorithm analysis were conducted on pivotal model genes to further elucidate their potential mechanisms in MG-associated DCM.
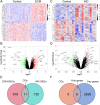
Results: In our analysis of both DCM and MG datasets, we identified 2641 critical module genes and 11 differentially expressed genes shared between the two conditions. Enrichment analysis disclosed that these 11 genes primarily pertain to inflammation and immune regulation. Connectivity map (CMAP) analysis pinpointed SB-216763 as a potential drug for DCM treatment. The results from machine learning indicated the substantial diagnostic value of midline 1 interacting protein1 (MID1IP1) and PI3K-interacting protein 1 (PIK3IP1) in MG-associated DCM. These two hub genes were chosen as candidate biomarkers and employed to formulate a diagnostic nomogram with optimal diagnostic performance through machine learning. Simultaneously, single-gene GSEA results and immune cell infiltration analysis unveiled immune dysregulation in both DCM and MG, with MID1IP1 and PIK3IP1 showing significant associations with invasive immune cells.
Conclusion: We have elucidated the inflammatory and immune pathways associated with MG-related DCM and formulated a diagnostic nomogram for DCM utilizing MID1IP1/PIK3IP1. This contribution offers novel insights for prospective diagnostic approaches and therapeutic interventions in the context of MG coexisting with DCM.
Keywords: Diagnostic value; Dilated cardiomyopathy; Immune cell infiltration; Machine learning algorithms; Myasthenia gravis.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures







References
-
- Querol L., Illa I. Myasthenia gravis and the neuromuscular junction. Curr. Opin. Neurol. 2013;26(5):459–465. - PubMed
-
- Japp A.G., et al. The diagnosis and evaluation of dilated cardiomyopathy. J. Am. Coll. Cardiol. 2016;67(25):2996–3010. - PubMed
-
- Namba T., Brunner N.G., Grob D. Idiopathic giant cell polymyositis. Report of a case and review of the syndrome. Arch. Neurol. 1974;31(1):27–30. - PubMed
-
- de Jongste M.J., Oosterhuis H.J., Lie K.I. Intractable ventricular tachycardia in a patient with giant cell myocarditis, thymoma and myasthenia gravis. Int. J. Cardiol. 1986;13(3):374–378. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

